

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
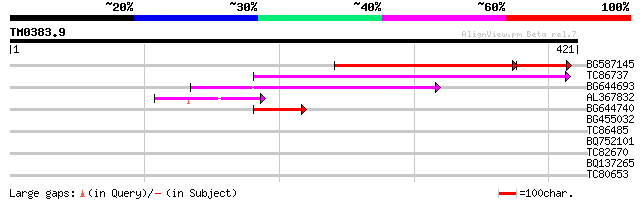
Query= TM0383.9
(421 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 152 2e-46
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 131 4e-31
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 86 4e-17
AL367832 weakly similar to GP|14091845|gb Putative retroelement ... 59 3e-09
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 49 4e-06
BG455032 weakly similar to GP|10440622|gb| putative NBS-LRR type... 30 1.7
TC86485 similar to GP|23315211|gb|AAN20392.1 Sequence 405 from p... 30 2.3
BQ752101 similar to PIR|S57177|S57 branched-chain-amino-acid tra... 29 3.9
TC82670 weakly similar to GP|18389232|gb|AAL67059.1 unknown prot... 28 5.1
BQ137265 similar to GP|21751020|dbj unnamed protein product {Hom... 28 8.7
TC80653 similar to GP|20257495|gb|AAM15917.1 purple acid phospha... 28 8.7
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 152 bits (385), Expect(2) = 2e-46
Identities = 70/136 (51%), Positives = 100/136 (73%)
Frame = +2
Query: 242 MHPSDEDKTAFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVK 301
MHP D +KTAF+T R YCY+ MPFGLKNAG+TYQRL++R+F ++G MEVY+DDM+VK
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 302 SVLGSSHHEDLTKAFARLRKHNMRLNPEKCSFGIQGGKFLGFMITTRGIEINLDKCKAIQ 361
S+ + H L + F L ++ M+LNP KC+FG+ G+FLG+++T +GIE+N + AI
Sbjct: 182 SLRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAIL 361
Query: 362 EMKSPGSVKEVQHLIG 377
++ SP + +EVQ L G
Sbjct: 362 DLPSPKNSREVQRLTG 409
Score = 51.2 bits (121), Expect(2) = 2e-46
Identities = 23/41 (56%), Positives = 29/41 (70%)
Frame = +3
Query: 377 GRIAALSRFLPHSGDKSAPFFKCLKKNAAFEWNSECEEAFK 417
GRIAAL+RF+ S DK PF+K L N F W+ +CEEAF+
Sbjct: 408 GRIAALNRFISRSTDKCLPFYKLLCGNKRFVWDEKCEEAFE 530
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 131 bits (330), Expect = 4e-31
Identities = 71/237 (29%), Positives = 124/237 (51%), Gaps = 2/237 (0%)
Frame = +1
Query: 182 VVMVKKANGKWRMCVDYTDLNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYHQIK 241
V+ V+K G R CVDY LN K+ YPLP I + + +G + +D + +H+++
Sbjct: 583 VLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETLRRVAGARWFTKLDVVAAFHKMR 762
Query: 242 MHPSDEDKTAFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVK 301
+ D++KTAF T + + PFGL A AT+QR +++ + + Y+DD+++
Sbjct: 763 IKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYINKTLHEFLDDFVTAYIDDVLIY 942
Query: 302 SVLGSSHHE-DLTKAFARLRKHNMRLNPEKCSFGIQGGKFLGFMITT-RGIEINLDKCKA 359
+ HE + + RL + L+P+KC F + K++GF++T +G+ + K A
Sbjct: 943 TTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVTTVKYVGFILTAGKGVSCDPLKLAA 1122
Query: 360 IQEMKSPGSVKEVQHLIGRIAALSRFLPHSGDKSAPFFKCLKKNAAFEWNSECEEAF 416
I++ PGSVK + +G F+P + + P + +K+ F W +E E AF
Sbjct: 1123IRDWLPPGSVKGARSFLGFCNYYKDFIPGYSEITEPLTRLTRKDFPFRWGAEQEAAF 1293
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 85.5 bits (210), Expect = 4e-17
Identities = 58/186 (31%), Positives = 90/186 (48%)
Frame = +2
Query: 135 LNPGIEPVTQTRRQMGDVKEKAIHQEVNKLLAADFIREIKYPTWMTNVVMVKKANGKWRM 194
L P + P+ ++ +K K + ++ LL FI+ YP + V+ +KK +G RM
Sbjct: 53 LLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVV-VLFLKKKDGFLRM 229
Query: 195 CVDYTDLNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYHQIKMHPSDEDKTAFMT 254
+DY LN K YPLP ID+L D G++ ID G HQ ++ D KTAF
Sbjct: 230 SIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVIGEDVPKTAFRI 409
Query: 255 ARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSSHHEDLTK 314
+Y M FG N + LM+RVF+ + + V+ +D+++ S + H L
Sbjct: 410 RYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSKNENEHENHLRL 589
Query: 315 AFARLR 320
A L+
Sbjct: 590 ALKVLK 607
>AL367832 weakly similar to GP|14091845|gb Putative retroelement {Oryza
sativa}, partial (2%)
Length = 384
Score = 59.3 bits (142), Expect = 3e-09
Identities = 35/86 (40%), Positives = 50/86 (57%), Gaps = 3/86 (3%)
Frame = -1
Query: 108 LLGENLDLFAWSHKDMPGIDPNFI--CLALNPGIEPVT-QTRRQMGDVKEKAIHQEVNKL 164
LL E LD+FA S++DMPG+DP + + P PV + RR D+ K I EV K
Sbjct: 255 LLREYLDIFACSYEDMPGLDPKIVEHRIPTKPECPPVR*KLRRTHPDMALK-IKSEVQKQ 79
Query: 165 LAADFIREIKYPTWMTNVVMVKKANG 190
+ A F+ ++YP W+ N+V V K +G
Sbjct: 78 IDAGFLMTVEYPEWVANIVPVPKKDG 1
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 48.9 bits (115), Expect = 4e-06
Identities = 20/39 (51%), Positives = 26/39 (66%)
Frame = -1
Query: 182 VVMVKKANGKWRMCVDYTDLNKACPKNSYPLPSIDKLVD 220
++ V+K +G +RMC+DY NK KN YPLP ID L D
Sbjct: 232 LLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFD 116
>BG455032 weakly similar to GP|10440622|gb| putative NBS-LRR type resistance
protein 3' partial {Oryza sativa}, partial (3%)
Length = 666
Score = 30.0 bits (66), Expect = 1.7
Identities = 19/64 (29%), Positives = 31/64 (47%), Gaps = 1/64 (1%)
Frame = +1
Query: 184 MVKKANGKWRMCVDYTDLNKACPKNSYP-LPSIDKLVDGASGNELMSLIDAYSGYHQIKM 242
++K G+ C+ D+ K CPK P LPS+ L NEL+ I + G Q+ +
Sbjct: 64 LLKVERGEMFPCLSSLDIWK-CPKLGLPCLPSLKDLGVDGRNNELLRSISTFRGLTQLTL 240
Query: 243 HPSD 246
+ +
Sbjct: 241 NSGE 252
>TC86485 similar to GP|23315211|gb|AAN20392.1 Sequence 405 from patent US
6410718, partial (90%)
Length = 1927
Score = 29.6 bits (65), Expect = 2.3
Identities = 14/40 (35%), Positives = 21/40 (52%)
Frame = +3
Query: 363 MKSPGSVKEVQHLIGRIAALSRFLPHSGDKSAPFFKCLKK 402
MKSP +K+VQ + + LSR + S + + KC K
Sbjct: 1131 MKSPEDLKKVQQELAEVVGLSRQVEESDFEKLTYLKCALK 1250
>BQ752101 similar to PIR|S57177|S57 branched-chain-amino-acid transaminase
(EC 2.6.1.42) BAT2 cytosolic - yeast, partial (46%)
Length = 720
Score = 28.9 bits (63), Expect = 3.9
Identities = 23/65 (35%), Positives = 30/65 (45%)
Frame = -2
Query: 49 QRGRPVREKERLGGTLM*RGRCAGREPRHKGRGTSQQAHADRGDQGAEIWREDTQDRDML 108
+RGRP R R L R R R+P H Q H R D+G +WR+ QD +
Sbjct: 257 RRGRPGRPSHR---GLWYRHR-RHRQP-HSHHCLEGQIHQLRSDRGRGVWRDCPQDEGLD 93
Query: 109 LGENL 113
GE +
Sbjct: 92 *GETV 78
>TC82670 weakly similar to GP|18389232|gb|AAL67059.1 unknown protein
{Arabidopsis thaliana}, partial (18%)
Length = 716
Score = 28.5 bits (62), Expect = 5.1
Identities = 13/31 (41%), Positives = 15/31 (47%)
Frame = +1
Query: 199 TDLNKACPKNSYPLPSIDKLVDGASGNELMS 229
T L + C KNSYPLP +G N S
Sbjct: 52 TKLQELCQKNSYPLPEYQTTREGPPHNSRFS 144
>BQ137265 similar to GP|21751020|dbj unnamed protein product {Homo
sapiens}, partial (9%)
Length = 1112
Score = 27.7 bits (60), Expect = 8.7
Identities = 20/52 (38%), Positives = 22/52 (41%)
Frame = +2
Query: 29 AEWTNWESGCGPKKGEGVLLQRGRPVREKERLGGTLM*RGRCAGREPRHKGR 80
A+ T W G GV L + G T RGRC GRE R KGR
Sbjct: 11 AQRTRWIPQAAKNSGPGVRLA--------QITGATAAARGRCEGRE-RSKGR 139
>TC80653 similar to GP|20257495|gb|AAM15917.1 purple acid phosphatase
{Arabidopsis thaliana}, partial (67%)
Length = 1661
Score = 27.7 bits (60), Expect = 8.7
Identities = 12/29 (41%), Positives = 20/29 (68%)
Frame = -2
Query: 216 DKLVDGASGNELMSLIDAYSGYHQIKMHP 244
++LV AS N L S + + S +HQ+++HP
Sbjct: 1630 EQLVGLASLNMLSSCLSSLSRFHQLQLHP 1544
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,500,667
Number of Sequences: 36976
Number of extensions: 179098
Number of successful extensions: 756
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 751
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 753
length of query: 421
length of database: 9,014,727
effective HSP length: 99
effective length of query: 322
effective length of database: 5,354,103
effective search space: 1724021166
effective search space used: 1724021166
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0383.9


