

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
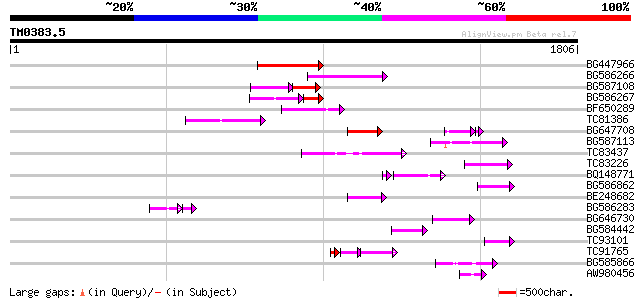
Query= TM0383.5
(1806 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [impo... 223 4e-58
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 213 7e-55
BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - ... 95 3e-36
BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At... 105 1e-34
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 135 2e-31
TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR re... 126 9e-29
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 117 3e-26
BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At... 115 2e-25
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 112 1e-24
TC83226 weakly similar to PIR|G86419|G86419 probable reverse tra... 96 1e-19
BQ148771 90 7e-18
BG586862 84 6e-16
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 80 7e-15
BG586283 similar to GP|7267666|em RNA-directed DNA polymerase-li... 59 1e-13
BG646730 75 3e-13
BG584442 74 7e-13
TC93101 68 3e-11
TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative ret... 64 6e-11
BG585866 62 2e-09
AW980456 61 4e-09
>BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 687
Score = 223 bits (569), Expect = 4e-58
Identities = 109/210 (51%), Positives = 145/210 (68%)
Frame = +2
Query: 790 WLAHGDRNTSFFHKKASQRKDRNSIARITDDRGRAWTEEEDIEEVLTKYFEDLFKATSPD 849
WL GD+NT FFH KASQR+ N I ++ D+ G EE++E +L YF +LF +++P
Sbjct: 5 WLKDGDKNTKFFHSKASQRRKVNEIKKLKDETGNWCKGEENVERLLITYFNNLFTSSNPT 184
Query: 850 DCEIVTNLVHGRITNEHKEIMEEPFTEEEVREALKKMHPTKAHGADGTPALFFQKFWSIV 909
E +V G++++EH E+ FTEEEV EA+ +MHP KA G DG PALFFQK+W IV
Sbjct: 185 AIEETCEVVKGKLSHEHIVWCEKEFTEEEVLEAINQMHPVKAPGPDGLPALFFQKYWHIV 364
Query: 910 GYDVTAQVLSILNDGADATHINQTLIALIPKIKKPSMAKDYRPISL*NVIFKLVTKTIAN 969
G +V VL +LN+ + +N+T I LIPK K P+ KDYRPISL NV+ K++TK IAN
Sbjct: 365 GKEVQQMVLQVLNNSMETEELNKTFIVLIPKGKNPNTPKDYRPISLCNVVMKIITKVIAN 544
Query: 970 RLKSFLQDIVEETQSAFVPGRLITDNAIIA 999
R+K L D+++ QSAFV GRLITDNA+IA
Sbjct: 545 RVKQTLPDVIDVEQSAFVQGRLITDNALIA 634
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 213 bits (541), Expect = 7e-55
Identities = 107/255 (41%), Positives = 153/255 (59%)
Frame = -3
Query: 949 DYRPISL*NVIFKLVTKTIANRLKSFLQDIVEETQSAFVPGRLITDNAIIAHECFHYMRR 1008
+YR I+ N +K++ K ++ R++ L+ I+ +QSAFVPGR I+DN +I H+ HY+R+
Sbjct: 772 EYRTIAPCNTQYKIIAKILSKRMQPLLRSIISPSQSAFVPGRAISDNVLITHKILHYLRQ 593
Query: 1009 KTRGVKGVMALKTDMSKAYDRVEWNFLKSTLEQMGFTQRWTNLIMNCVSTVSFQVLLNGQ 1068
MA+KTDM+KAYDR+ WNFL+ L ++GF W + IM CVSTVS+ L+NG
Sbjct: 592 SGAKKHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFHGIWISWIMECVSTVSYSFLINGG 413
Query: 1069 PCRSFKPERGLRQGDPLSPYLFILCAEVFSSMIKAKVAAGVLHGIKITPRAPMISHLFFA 1128
P P RGLRQGDPLSPYLFILC EV S + + + G L G+K+ P I+HL FA
Sbjct: 412 PQGRVLPSRGLRQGDPLSPYLFILCTEVLSGLCQQALRKGTLPGVKVARNCPPINHLLFA 233
Query: 1129 DDSILLARATKEEAMVLHDIIKGYEAFSGQRINFDKSELSTSRNVPSTRVHELEQLLGVK 1188
DD++ ++ +L I+ Y A SG+ IN KS ++ S + ++ L +
Sbjct: 232 DDTMFFGKSNASSCAILLSIMDKYRAASGRCIN*TKSAITFSSKTSQAIIDRVKGELKIA 53
Query: 1189 AVEKHGKYLGLPTIV 1203
GKYLG I+
Sbjct: 52 KEGGTGKYLGYRNIL 8
>BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - Arabidopsis
thaliana, partial (17%)
Length = 677
Score = 95.1 bits (235), Expect(2) = 3e-36
Identities = 54/135 (40%), Positives = 72/135 (53%)
Frame = +2
Query: 768 EKELENLLKLEEIKWSQRSRATWLAHGDRNTSFFHKKASQRKDRNSIARITDDRGRAWTE 827
+++L+ K EE W Q+SR W GD N F+H QR RN I + D G TE
Sbjct: 11 KEKLQEAYKDEEDYWQQKSRNMWHISGDLNKKFYHALTKQRHARNRIVGLYDYDGNWITE 190
Query: 828 EEDIEEVLTKYFEDLFKATSPDDCEIVTNLVHGRITNEHKEIMEEPFTEEEVREALKKMH 887
E+ +E+V YFEDLF+ T+P + + + IT + + + TEEEVR AL MH
Sbjct: 191 EQGVEKVAVDYFEDLFQRTTPTGFDGFLDEITSSITPQMNQRLLRLATEEEVRLALFIMH 370
Query: 888 PTKAHGADGTPALFF 902
P KA G DG L F
Sbjct: 371 PEKAPGPDGMTTLLF 415
Score = 77.4 bits (189), Expect(2) = 3e-36
Identities = 41/89 (46%), Positives = 54/89 (60%)
Frame = +3
Query: 901 FFQKFWSIVGYDVTAQVLSILNDGADATHINQTLIALIPKIKKPSMAKDYRPISL*NVIF 960
FFQ W I+ D+ V S L G T +N T I LIPK K+P+ + RPISL NV +
Sbjct: 411 FFQHSWHIIKMDLLKMVNSFLASGKLDTRLNTTNICLIPKKKRPTRMTELRPISLCNVGY 590
Query: 961 KLVTKTIANRLKSFLQDIVEETQSAFVPG 989
K+++K + RLK L ++ ETQSAFV G
Sbjct: 591 KIISKVLCQRLKVCLPSLISETQSAFVHG 677
>BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (16%)
Length = 794
Score = 105 bits (263), Expect(2) = 1e-34
Identities = 64/179 (35%), Positives = 88/179 (48%), Gaps = 6/179 (3%)
Frame = +3
Query: 763 KGKEAE---KELENLLKLEEIKWSQRSRATWLAHGDRNTSFFHKKASQRKDRNSIARITD 819
KG+E EL EEI W Q+SR WL GDRNT FFH R+ +N I + D
Sbjct: 69 KGEELSLLRSELNEEYHNEEIFWMQKSRLNWLRSGDRNTKFFHAVTKNRRAQNRILSLID 248
Query: 820 DRGRAWTEEEDIEEVLTKYFEDLFKATSPDDCEIVT---NLVHGRITNEHKEIMEEPFTE 876
D + W EED+ + +F+ L+ S +D I N + +T E + +
Sbjct: 249 DDDKEWFVEEDLGRLADSHFKLLY---SSEDVGITLEDWNSIPAIVTEEQNAQLMAQISR 419
Query: 877 EEVREALKKMHPTKAHGADGTPALFFQKFWSIVGYDVTAQVLSILNDGADATHINQTLI 935
EEVREA+ ++P K G DG FFQ+FW +G D+T+ L G IN+T I
Sbjct: 420 EEVREAVFDINPHKCPGPDGMNVFFFQQFWDTMGDDLTSMAQEFLRTGKLEEGINKTNI 596
Score = 61.2 bits (147), Expect(2) = 1e-34
Identities = 31/64 (48%), Positives = 45/64 (69%)
Frame = +1
Query: 937 LIPKIKKPSMAKDYRPISL*NVIFKLVTKTIANRLKSFLQDIVEETQSAFVPGRLITDNA 996
L+PK + ++RPISL NV +K+V+K ++ RLKS L I+ ETQ+AF +LI+DN
Sbjct: 601 LVPKKLEAKRLVEFRPISLCNVAYKIVSKVLSKRLKSVLPWIITETQAAFGRRQLISDNI 780
Query: 997 IIAH 1000
+IAH
Sbjct: 781 LIAH 792
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 135 bits (339), Expect = 2e-31
Identities = 72/201 (35%), Positives = 114/201 (55%)
Frame = +3
Query: 867 KEIMEEPFTEEEVREALKKMHPTKAHGADGTPALFFQKFWSIVGYDVTAQVLSILNDGAD 926
++++ FT EV+ AL M +KA G DG FF+ W+I+G V +L G
Sbjct: 3 QDLLCSEFTAVEVKNALFSMDSSKAPGIDGYNVHFFKCSWNIIGDSVIDAILDFFKTGFM 182
Query: 927 ATHINQTLIALIPKIKKPSMAKDYRPISL*NVIFKLVTKTIANRLKSFLQDIVEETQSAF 986
IN T + L+PK + K++RPI+ +VI+K+++K + +R++ L +V E QSAF
Sbjct: 183 PKIINCTYVTLLPKEVNVTSVKNFRPIACCSVIYKIISKILTSRMQGVLNSVVSENQSAF 362
Query: 987 VPGRLITDNAIIAHECFHYMRRKTRGVKGVMALKTDMSKAYDRVEWNFLKSTLEQMGFTQ 1046
V GR+I DN I++HE RK G+ +K D+ KAYD EW F+K + ++GF
Sbjct: 363 VKGRVIFDNIILSHELVKSYSRK--GISPRCMVKIDLXKAYDSXEWPFIKHLMLELGFPY 536
Query: 1047 RWTNLIMNCVSTVSFQVLLNG 1067
++ N +M ++T S+ NG
Sbjct: 537 KFVNWVMAXLTTASYTFNXNG 599
>TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR retroelement
reverse transcriptase {Oryza sativa (japonica
cultivar-group)}, partial (1%)
Length = 798
Score = 126 bits (316), Expect = 9e-29
Identities = 75/264 (28%), Positives = 126/264 (47%), Gaps = 9/264 (3%)
Frame = -1
Query: 561 WELIKKLTPDPEVPWLCLGDFNEVLKSSEKTGGNPINFVGAQAFQEAMDFCGLRDMGFSG 620
W I + ++ W C+GDFN +L S E G + + FQ+ D L + G
Sbjct: 783 WSAISNIQTQHKLSWCCIGDFNTILGSHEHQGSHTPARLPMLDFQQWSDVNNLFHLPTRG 604
Query: 621 FQFTWSNMRPTPNTIQERLDRALVNECWTEHWPHTHVSHLDRFNSDHNPIYVIFLKAKRQ 680
FTW+N R N ++RLDR++VN+ ++ L + SDH PI + + Q
Sbjct: 603 SAFTWTNGRRGRNNTRKRLDRSIVNQLMIDNCESLSACTLTKLRSDHFPI---LFELQTQ 433
Query: 681 KKKKIRLYRFEEMWLQEEDCRHVIHHSWPRGGRN-----ITSKLGNVGANLTEWAEATFG 735
+ ++F +MW DC ++I W + KL N+ L W + TFG
Sbjct: 432 NIQFSSSFKFMKMWSAHPDCINIIKQCWANQVVGCPMFVLNQKLKNLKEVLKVWNKNTFG 253
Query: 736 NIQKRI----KSQLGIIKRIQAAEQTEENLKKGKEAEKELENLLKLEEIKWSQRSRATWL 791
N+ ++ K I +I + ++ + + K A+ LE+ L +EE+ W ++S+ W
Sbjct: 252 NVHSQVDNAYKELDDIQVKIDSIGYSDVLMDQEKAAQLNLESALNIEEVFWHEKSKVNWH 73
Query: 792 AHGDRNTSFFHKKASQRKDRNSIA 815
GDRNT+FFH+ A ++ + I+
Sbjct: 72 CEGDRNTAFFHRVAKIKRTSSLIS 1
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 117 bits (294), Expect = 3e-26
Identities = 57/114 (50%), Positives = 78/114 (68%)
Frame = +1
Query: 1075 PERGLRQGDPLSPYLFILCAEVFSSMIKAKVAAGVLHGIKITPRAPMISHLFFADDSILL 1134
PE+GLRQGDPLSPYLFILCA V S ++K + LHGI++ P I+HL FADDS+L
Sbjct: 4 PEKGLRQGDPLSPYLFILCANVLSGLLKREGNKQNLHGIQVARSDPKITHLLFADDSLLF 183
Query: 1135 ARATKEEAMVLHDIIKGYEAFSGQRINFDKSELSTSRNVPSTRVHELEQLLGVK 1188
ARA EA + ++ Y++ SGQ +NF+KSE+S S+NVP+ + Q + +K
Sbjct: 184 ARANLTEAATIMQVLHSYQSASGQLVNFEKSEVSYSQNVPNQEKEMICQQIAIK 345
Score = 48.5 bits (114), Expect(2) = 4e-06
Identities = 30/99 (30%), Positives = 48/99 (48%)
Frame = +3
Query: 1385 WLPNSSSGRVTTPRNANEGPQRVCELILAGRGMWNESLIKATFSGYDASKILQIPLRGTH 1444
W+P+ ++ N G V ELI WN LI +F+ Y A +I++IPL
Sbjct: 348 WIPSKREFKL----NHVSGSLSVDELIDYDTKQWNRDLIFHSFNNYAAHQIIKIPLSMRQ 515
Query: 1445 AKDQLIWKDSGVGQYSVKAGYRIQRSQNNTHPATGPSGT 1483
+D++IW G YSV++ + +N ++ A S T
Sbjct: 516 PEDKIIWHWEKDGIYSVRSAHHALCDENFSNQAESSSAT 632
Score = 21.9 bits (45), Expect(2) = 4e-06
Identities = 6/24 (25%), Positives = 10/24 (41%)
Frame = +2
Query: 1484 NHSNLWKKVWKVKTLPRCNEIIWR 1507
N+ +WK +W +WR
Sbjct: 629 NNPVIWKAIWNTPASRSVQNFLWR 700
>BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (10%)
Length = 767
Score = 115 bits (287), Expect = 2e-25
Identities = 80/261 (30%), Positives = 116/261 (43%), Gaps = 15/261 (5%)
Frame = -3
Query: 1341 PLEANRGTNSSYLWKSVHAAREVIQKGSIWKVGTGEKIHIWNDQWL---------PNSSS 1391
PL A G+ +SY W+S+H+A+ +I++G+ +G GE +IW + PN S
Sbjct: 765 PLNAPLGSWASYAWRSIHSAQHLIKQGAKVIIGNGENTNIWEREMAWKLTCVTNHPNKHS 586
Query: 1392 GRV--TTPRNANEGPQRVCELILAGRGMWNESLIKATFSGYDASKILQIPLRGTHAKDQL 1449
R EG C R N +LI + F KIL I +G +D
Sbjct: 585 SRAY*APTLYGYEG----CRSDDPMRRERNANLINSIFPEGTRRKILSIHPQGPIGEDSY 418
Query: 1450 IWKDSGVGQYSVKAGYRIQRSQNNTHPATGPSGT----NHSNLWKKVWKVKTLPRCNEII 1505
W+ S G YSVK+GY +Q N A GT + +L+++VWK T P+ +
Sbjct: 417 SWEYSKSGHYSVKSGYYVQ---TNIIAAANQRGTVDQPSLDDLYQRVWKYNTSPKVRHFL 247
Query: 1506 WRACKGVLPVRMNLKRRGMDLDTKCPCCGEEDETTTHALLTCSSIRRLWFASHLNLHLNE 1565
WR LP N++ R + D C CG E ET H L C R +W S ++
Sbjct: 246 WRCISNSLPTAANMRSRHISKDGSCSRCGMESETVNHILFQCPYARLIWATSPIHAPPYG 67
Query: 1566 ELPDNFAGWLEQVLELPNEET 1586
+ D+ L +VL L T
Sbjct: 66 IMYDSLYSNLHRVLSLQERYT 4
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 112 bits (281), Expect = 1e-24
Identities = 94/336 (27%), Positives = 149/336 (43%), Gaps = 2/336 (0%)
Frame = +2
Query: 930 INQTLIALIPKIKKPSMAKDYRPISL*NVIFKLVTKTIANRLKSFLQDIVEETQSAFVPG 989
IN T IALIPK+ P D+RPISL ++K++ K +ANRL+ + ++ + QSAFV
Sbjct: 50 INSTFIALIPKVDNPQRLNDFRPISLVGSLYKILGKLLANRLRVVIGSVISDAQSAFVKN 229
Query: 990 RLITDNAIIAHECFHYMRRKTRGVKGVMA--LKTDMSKAYDRVEWNFLKSTLEQMGFTQR 1047
R I + + R R + + LK ++ + + F M F
Sbjct: 230 RQILEMVFL*QMRLWMRLRN*RKIFCCLRWILKRLITLSIGLIWILF*VG----MSFLVL 397
Query: 1048 WTNLIMNCVSTVSFQVLLNGQPCRSFKPERGLRQGDPLSPYLFILCAEVFSSMIKAKVAA 1107
W I CVST + VL+NG P +L + + + + +
Sbjct: 398 WRKWIKECVSTATTSVLVNGSPTN-------------------VLMKSLVQTQLFTRYSF 520
Query: 1108 GVLHGIKITPRAPMISHLFFADDSILLARATKEEAMVLHDIIKGYEAFSGQRINFDKSEL 1167
GV++ + ++SHL FA+D++LL L + + A SG ++NF KS L
Sbjct: 521 GVVNPV-------VVSHLQFANDTLLLETKNWANIRALRAALVIF*AMSGLKVNFHKSGL 679
Query: 1168 STSRNVPSTRVHELEQLLGVKAVEKHGKYLGLPTIVGRSKTQVFSFVRERIWKKLKGWKE 1227
PS + E +L K + YLG+P + + + RI +L GW
Sbjct: 680 VCVNIAPSW-LSEAASVLSWKVGKVPFLYLGMPIEGNSRRLSFWEPIVNRIKARLTGWNS 856
Query: 1228 KALSRSGREVLIKSIAQAIPSYVMGLFLLPSTLCNQ 1263
+ LS GR VL+KS+ ++ Y LPS+ +Q
Sbjct: 857 RFLSFGGRLVLLKSVLTSLSVYA-----LPSSKLHQ 949
>TC83226 weakly similar to PIR|G86419|G86419 probable reverse transcriptase
100033-105622 [imported] - Arabidopsis thaliana, partial
(2%)
Length = 885
Score = 95.9 bits (237), Expect = 1e-19
Identities = 46/153 (30%), Positives = 80/153 (52%)
Frame = +3
Query: 1450 IWKDSGVGQYSVKAGYRIQRSQNNTHPATGPSGTNHSNLWKKVWKVKTLPRCNEIIWRAC 1509
+W + G YSVK+GY R+ + ++ + +WKK+W + T+PR ++WR
Sbjct: 3 MWMHNPTGIYSVKSGYNTLRTWQTQQINNTSTSSDETLIWKKIWSLHTIPRHKVLLWRIL 182
Query: 1510 KGVLPVRMNLKRRGMDLDTKCPCCGEEDETTTHALLTCSSIRRLWFASHLNLHLNEELPD 1569
LPVR +L++RG+ CP C + ET TH ++C +R+WF S+L ++ +
Sbjct: 183 NDSLPVRSSLRKRGIQCYPLCPRCHSKTETITHLFMSCPLSKRVWFGSNLCINFDNLPNP 362
Query: 1570 NFAGWLEQVLELPNEETMADLFNLIWVIWKRRN 1602
NF L + + +E + +I+ +W RN
Sbjct: 363 NFIN*LYEAIL*KDECITI*IAAIIYNLWHARN 461
>BQ148771
Length = 680
Score = 89.7 bits (221), Expect(2) = 7e-18
Identities = 54/173 (31%), Positives = 88/173 (50%), Gaps = 7/173 (4%)
Frame = -3
Query: 1222 LKGWKEKALSRSGREVLIKSIAQAIPSYVMGLFLLPSTLCNQVERMINSFYWGGSADSRK 1281
L WK LS + R L KS+ +A+P Y M ++P +++++ F WG + SR+
Sbjct: 573 LANWKANHLSLARRVTLAKSVIEAVPLYPMMTTIIPKACIEEIQKLQRKFVWGDTEVSRR 394
Query: 1282 LNWIRWDTLATPKNMGGLGFRSFHSFNKALLAKQWWRLINRENSLMVQVIKARYYPKSTP 1341
+ + W+T++ PK + GLG R NKA + K W + + NSL +V++ + Y +S
Sbjct: 393 YHAVGWETMSKPKTIYGLGLRRLDVMNKACIMKLGWSIYSGSNSLCTEVMRGK-YQRSES 217
Query: 1342 LE---ANRGTNSSYLWKSVHAAREVIQKGSIWKVGTGEKIHIWN----DQWLP 1387
LE + T+SS LWK++ I++ + G WN QWLP
Sbjct: 216 LEEIFLEKPTDSS-LWKALVKLWPEIERNLVDSNGN------WNWEKLKQWLP 79
Score = 20.8 bits (42), Expect(2) = 7e-18
Identities = 9/29 (31%), Positives = 13/29 (44%)
Frame = -2
Query: 1186 GVKAVEKHGKYLGLPTIVGRSKTQVFSFV 1214
G + GKY G+P K +FS +
Sbjct: 676 GFRXATSLGKYXGVPC*EEHQKEPIFSIL 590
>BG586862
Length = 804
Score = 83.6 bits (205), Expect = 6e-16
Identities = 39/117 (33%), Positives = 60/117 (50%)
Frame = -1
Query: 1490 KKVWKVKTLPRCNEIIWRACKGVLPVRMNLKRRGMDLDTKCPCCGEEDETTTHALLTCSS 1549
+KVW +KT+PR +WR LPV+ L +RG+ CP C + ET H L C
Sbjct: 660 EKVWGIKTIPRHKSFLWRLLHNALPVKDELHKRGIRCSLLCPRCESKIETVQHLFLNCEV 481
Query: 1550 IRRLWFASHLNLHLNEELPDNFAGWLEQVLELPNEETMADLFNLIWVIWKRRNAWIF 1606
++ WF S L ++ + +F W+ + +EET+ L L++ IW RN +F
Sbjct: 480 TQKEWFGSQLGINFHSSGVLHFHDWITNFILKNDEETIIALTALLYSIWHARNQKVF 310
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 80.1 bits (196), Expect = 7e-15
Identities = 44/125 (35%), Positives = 68/125 (54%)
Frame = +3
Query: 1076 ERGLRQGDPLSPYLFILCAEVFSSMIKAKVAAGVLHGIKITPRAPMISHLFFADDSILLA 1135
+RGL+QGDPL+P+LF+L AE S ++K V + G + +SHL +ADD++ +
Sbjct: 21 QRGLKQGDPLAPFLFLLVAEGISGLMKNAVNRNLFQGFDVKRGGTRVSHLQYADDTLCIG 200
Query: 1136 RATKEEAMVLHDIIKGYEAFSGQRINFDKSELSTSRNVPSTRVHELEQLLGVKAVEKHGK 1195
T + L +++G+E SG ++NF KS L NVP + + L +
Sbjct: 201 MPTVDNLWTLKALLQGFEMASGLKVNFHKSSL-IGINVPRDFMEAACRFLNCREESIPFI 377
Query: 1196 YLGLP 1200
YLGLP
Sbjct: 378 YLGLP 392
>BG586283 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (7%)
Length = 774
Score = 58.9 bits (141), Expect(2) = 1e-13
Identities = 35/106 (33%), Positives = 55/106 (51%)
Frame = +2
Query: 444 WNCRGLGTPRAVGALRKLITMERPDLVFLMETRKKAFEIQKTKMPSGLVHSIGVDCNGEG 503
WNC+G+G+ V LR+ T + PD++FLMET++K ++ K + + V G
Sbjct: 218 WNCQGIGSDLTVRRLREFQTKDSPDIMFLMETKRKDEDVYKMYRGTEFTNHFTVPPVG-- 391
Query: 504 KSRGGGLACYWSSSLDVNIISLSSNHIQMEVTTTIIDSKMVVTGIY 549
GGLA W ++ V I++ S+N I +V D V+ IY
Sbjct: 392 --LSGGLALSWKDNVQVEILASSANVIDTKVNYN--DKSFFVSYIY 517
Score = 37.4 bits (85), Expect(2) = 1e-13
Identities = 15/44 (34%), Positives = 25/44 (56%)
Frame = +3
Query: 550 GHPEAPQKWRTWELIKKLTPDPEVPWLCLGDFNEVLKSSEKTGG 593
G P+ + + W+ + + + WL GDFN++L +SEK GG
Sbjct: 519 GVPQPEHRAKFWDELSTIGAQRDGAWLLTGDFNDLLDNSEKVGG 650
>BG646730
Length = 799
Score = 74.7 bits (182), Expect = 3e-13
Identities = 40/135 (29%), Positives = 62/135 (45%)
Frame = +1
Query: 1347 GTNSSYLWKSVHAAREVIQKGSIWKVGTGEKIHIWNDQWLPNSSSGRVTTPRNANEGPQR 1406
G SY W+S+ ++VI GS W + G+ + IW D WLPN + +V +P E
Sbjct: 388 GNQPSYAWRSMFNIKDVIDLGSRWSISNGQNVRIWKDDWLPNQTGFKVCSPMVDFEEATC 567
Query: 1407 VCELILAGRGMWNESLIKATFSGYDASKILQIPLRGTHAKDQLIWKDSGVGQYSVKAGYR 1466
+ ELI L++ FS ++A +IL IPL D+ I YSV++ +
Sbjct: 568 ISELIDTNTKSSKRDLVRNVFSAFEAKQILNIPLSWRLWPDRRILYWERDRNYSVRSAHH 747
Query: 1467 IQRSQNNTHPATGPS 1481
+ + P S
Sbjct: 748 LIKVATKDSPEVSSS 792
>BG584442
Length = 775
Score = 73.6 bits (179), Expect = 7e-13
Identities = 41/118 (34%), Positives = 70/118 (58%), Gaps = 1/118 (0%)
Frame = +1
Query: 1215 RERIWKKLKGWKEKALSRSGREVLIKSIAQAIPSYVMGLFLLPSTLCNQVERMINSFYWG 1274
R + KK+ + K LS+ EV+IK Q+I SYVM +FLL ++ +++E+++N+F W
Sbjct: 364 RTKFDKKINF*RNKCLSKVM*EVMIKYALQSISSYVMSIFLLLNSQVDEIEKIMNTFSWV 543
Query: 1275 GSADSRK-LNWIRWDTLATPKNMGGLGFRSFHSFNKALLAKQWWRLINRENSLMVQVI 1331
++RK ++W+ + L KN GG+GF F +FN +L KQ + +L ++ I
Sbjct: 544 HVGENRKGMHWMS*EKLFVHKNYGGMGFTDFTTFNIPMLGKQV*SFLLNRTTLFLEKI 717
>TC93101
Length = 675
Score = 68.2 bits (165), Expect = 3e-11
Identities = 32/94 (34%), Positives = 48/94 (51%)
Frame = -1
Query: 1513 LPVRMNLKRRGMDLDTKCPCCGEEDETTTHALLTCSSIRRLWFASHLNLHLNEELPDNFA 1572
LPVR L +RG++ CP C ETT H ++C +R+WF S L++ + NF+
Sbjct: 663 LPVRXELNKRGVNCPPLCPRCYFNLETTNHIFMSCERTQRVWFGSQLSIRFPDNSTINFS 484
Query: 1573 GWLEQVLELPNEETMADLFNLIWVIWKRRNAWIF 1606
WL + EE + + + + IW RN IF
Sbjct: 483 DWLFDAISNQTEEIIIKISAITYSIWHARNKAIF 382
>TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative retroelement
{Oryza sativa (japonica cultivar-group)}, partial (1%)
Length = 625
Score = 63.9 bits (154), Expect = 5e-10
Identities = 41/120 (34%), Positives = 65/120 (54%), Gaps = 1/120 (0%)
Frame = +3
Query: 1116 TPRAPMISH-LFFADDSILLARATKEEAMVLHDIIKGYEAFSGQRINFDKSELSTSRNVP 1174
TPR I L F + R + A ++ +I+ YE SG+ I+ KS + SRNVP
Sbjct: 249 TPRRGAIPMALLFEGAPLRSLRVEEHHAQIMKNILILYEEDSGKAISLRKS*IYCSRNVP 428
Query: 1175 STRVHELEQLLGVKAVEKHGKYLGLPTIVGRSKTQVFSFVRERIWKKLKGWKEKALSRSG 1234
+ +LGV+ + KYLGLP+++GR +T FS ++ +W+K K + E+ +S G
Sbjct: 429 DILKTSITYILGVQFMLGTCKYLGLPSMIGRDRTTTFSSIKGGVWQKNK-FLERHMSIQG 605
Score = 55.5 bits (132), Expect(2) = 6e-11
Identities = 29/69 (42%), Positives = 37/69 (53%)
Frame = +2
Query: 1055 CVSTVSFQVLLNGQPCRSFKPERGLRQGDPLSPYLFILCAEVFSSMIKAKVAAGVLHGIK 1114
CV + + VL+N P RGL+QGD LSPY+FI+C E S +I G HG
Sbjct: 104 CVESNDYYVLVNNDAVDPIIPSRGLQQGDHLSPYIFIICVEGLSFLIPHAKERGDTHGTS 283
Query: 1115 ITPRAPMIS 1123
I AP +S
Sbjct: 284 I*RGAPPVS 310
Score = 31.6 bits (70), Expect(2) = 6e-11
Identities = 12/26 (46%), Positives = 19/26 (72%)
Frame = +1
Query: 1023 MSKAYDRVEWNFLKSTLEQMGFTQRW 1048
+SK Y+RV+ ++LK + +MGF RW
Sbjct: 7 ISKVYNRVD*DYLKEIMIKMGFNNRW 84
>BG585866
Length = 828
Score = 62.0 bits (149), Expect = 2e-09
Identities = 49/203 (24%), Positives = 82/203 (40%), Gaps = 5/203 (2%)
Frame = +3
Query: 1357 VHAARE--VIQKGSIWKVGTGEKIHIWNDQWLPNSSSGRVTTPRNANEGPQRVCELILAG 1414
+H R+ V++ G W+ G+G W W G + ++ V ++ G
Sbjct: 78 IHIIRDKNVLKSGYTWRAGSGNS-SFWYTNWSSLGLLGTQAPFVDIHDLHLTVKDVFTTG 254
Query: 1415 RGMWNESLIKATFSGYDASKILQIPLRGTHAK--DQLIWKDSGVGQYSVKAGYRIQRSQN 1472
G +SL T D ++++ +A D IW + G Y+ K+GY SQ
Sbjct: 255 -GQHTQSLY--TILPTDIAEVINNTHLNFNASIGDAYIWPHNSNGVYTAKSGYSWILSQT 425
Query: 1473 NTHPATGPSGTNHSNL-WKKVWKVKTLPRCNEIIWRACKGVLPVRMNLKRRGMDLDTKCP 1531
T N++N W +W++K + +W AC +P L R M C
Sbjct: 426 ET--------VNYNNSSWSWIWRLKIPEKYKFFLWLACHNAVPTLSLLNHRNMVNSAICS 581
Query: 1532 CCGEEDETTTHALLTCSSIRRLW 1554
CGE +E+ H + C + +W
Sbjct: 582 RCGEHEESFFHCVRDCRFSKIIW 650
>AW980456
Length = 779
Score = 60.8 bits (146), Expect = 4e-09
Identities = 31/88 (35%), Positives = 44/88 (49%)
Frame = -2
Query: 1432 ASKILQIPLRGTHAKDQLIWKDSGVGQYSVKAGYRIQRSQNNTHPATGPSGTNHSNLWKK 1491
A KI+ PL D+LIWKD G+Y VK+ YR + S + W
Sbjct: 253 ADKIMSTPLISHA*LDRLIWKDEKHGKYYVKSAYRFCVEE-----LFDSSYLHRPGNWSG 89
Query: 1492 VWKVKTLPRCNEIIWRACKGVLPVRMNL 1519
+WK+K P+ ++WR C+G LP R+ L
Sbjct: 88 IWKLKVPPKVQNLVWRMCRGCLPTRIRL 5
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.333 0.144 0.484
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 63,686,798
Number of Sequences: 36976
Number of extensions: 993518
Number of successful extensions: 5790
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 4181
Number of HSP's successfully gapped in prelim test: 189
Number of HSP's that attempted gapping in prelim test: 1436
Number of HSP's gapped (non-prelim): 4610
length of query: 1806
length of database: 9,014,727
effective HSP length: 110
effective length of query: 1696
effective length of database: 4,947,367
effective search space: 8390734432
effective search space used: 8390734432
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0383.5


