

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
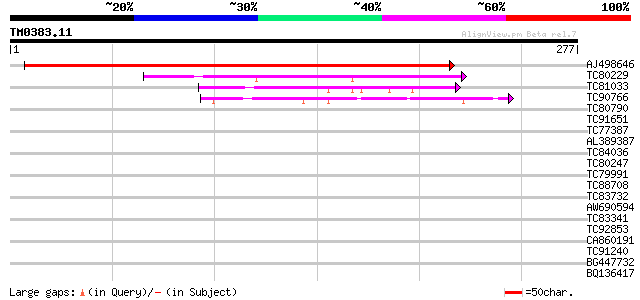
Query= TM0383.11
(277 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AJ498646 similar to PIR|E96759|E96 probable tumor-related protei... 368 e-102
TC80229 similar to PIR|H86215|H86215 protein T6D22.22 [imported]... 105 2e-23
TC81033 similar to PIR|T02654|T02654 hypothetical protein At2g26... 59 2e-09
TC90766 similar to PIR|T05491|T05491 hypothetical protein T19K4.... 42 3e-04
TC80790 similar to PIR|T38541|T38541 probable sucrose carrier - ... 37 0.011
TC91651 SP|P40389|UV22_SCHPO UV-induced protein uvi22. [Fission ... 35 0.041
TC77387 similar to GP|15983396|gb|AAL11566.1 At1g51440/F5D21_19 ... 32 0.20
AL389387 similar to GP|9759518|dbj| gene_id:MLN1.9~pir||T38261~s... 32 0.35
TC84036 GP|493598|gb|AAA89204.1|| major immediate early protein ... 30 1.0
TC80247 similar to PIR|C86476|C86476 protein F15O4.45 [imported]... 30 1.0
TC79991 similar to GP|10177307|dbj|BAB10568. contains similarity... 29 1.7
TC88708 29 1.7
TC83732 similar to GP|11762164|gb|AAG40360.1 AT4g22310 {Arabidop... 29 1.7
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 29 2.3
TC83341 weakly similar to GP|8978076|dbj|BAA98104.1 gene_id:K14A... 29 2.3
TC92853 similar to GP|20268748|gb|AAM14077.1 unknown protein {Ar... 29 2.3
CA860191 homologue to GP|19697342|gb hypothetical protein {Dicty... 29 2.3
TC91240 homologue to PIR|T48518|T48518 transcription factor like... 28 3.0
BG447732 homologue to GP|9837385|gb|A retinitis pigmentosa GTPas... 28 3.0
BQ136417 weakly similar to EGAD|146423|156 vitellogenin {Anolis ... 28 3.9
>AJ498646 similar to PIR|E96759|E96 probable tumor-related protein T9L24.46
[imported] - Arabidopsis thaliana, partial (50%)
Length = 665
Score = 368 bits (944), Expect = e-102
Identities = 184/210 (87%), Positives = 196/210 (92%)
Frame = +2
Query: 8 EEEEEEEEDAPPPMVKLGSYGGEVRLVVPGEESAAEETMLLWGIQQPTLSKPNAFVSQSS 67
E EEEEE+A MVKLGSYGG V LVVPGEESAAEETMLLWGIQQPTLSKPNAFV+QSS
Sbjct: 35 ERMEEEEENATAAMVKLGSYGGSVMLVVPGEESAAEETMLLWGIQQPTLSKPNAFVAQSS 214
Query: 68 LQLRLDSCGHSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLGTLVLQEKKIVE 127
LQLRLDSCGHSLSILQSPSSLG PGVTG+VMWDSG+VLGKFLEHSVD G LVLQ KKIVE
Sbjct: 215 LQLRLDSCGHSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVE 394
Query: 128 LGSGCGLVGCIAALLGGEVILTDLLDRLRLLRKNIETNMKHVSLRGSATATELTWGEDPD 187
LGSGCGLVGCIAALLGGEVILTDL DR+RLLRKNIETNMKH+SLRGS TATELTWG+DPD
Sbjct: 395 LGSGCGLVGCIAALLGGEVILTDLPDRMRLLRKNIETNMKHISLRGSITATELTWGDDPD 574
Query: 188 RELIDPTPDFVVGSDVVYSENAVVDLVETL 217
+ELIDPTPD+++GSDVVYSE AVVDL+ETL
Sbjct: 575 QELIDPTPDYILGSDVVYSEGAVVDLLETL 664
>TC80229 similar to PIR|H86215|H86215 protein T6D22.22 [imported] -
Arabidopsis thaliana, partial (19%)
Length = 697
Score = 105 bits (261), Expect = 2e-23
Identities = 58/166 (34%), Positives = 98/166 (58%), Gaps = 8/166 (4%)
Frame = +3
Query: 66 SSLQLRLDSCGHSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLGTLV---LQE 122
++ ++ L+ GH L Q P+S G +WD+ +V KFLE + G L+
Sbjct: 153 TTFEMPLEVLGHDLLFAQDPNSKHH----GTTIWDASLVFAKFLERNCRKGRFSPAKLKG 320
Query: 123 KKIVELGSGCGLVGCIAALLGGEVILTDLLDRLRLLRKNIETNM-----KHVSLRGSATA 177
K+++ELG+GCG+ G A+LG +VI+TD + L LL++N++ N+ K+ L GS
Sbjct: 321 KRVIELGAGCGVSGFAMAMLGCDVIVTDQKEVLPLLQRNVDRNISRVMQKNPELFGSIKV 500
Query: 178 TELTWGEDPDRELIDPTPDFVVGSDVVYSENAVVDLVETLGQLSGP 223
+EL WG++ + + P D+++G+DVVY E+ + L++ + LSGP
Sbjct: 501 SELQWGDESHIKAVGPPFDYIIGTDVVYVEHLLEPLLQKILALSGP 638
>TC81033 similar to PIR|T02654|T02654 hypothetical protein At2g26810
[imported] - Arabidopsis thaliana, partial (54%)
Length = 697
Score = 59.3 bits (142), Expect = 2e-09
Identities = 39/136 (28%), Positives = 74/136 (53%), Gaps = 8/136 (5%)
Frame = +3
Query: 93 VTGAVMWDSGVVLGKFLEHSVDLGTLVLQEKKIVELGSGCGLVGCIAALLGGEVILTDLL 152
+TG ++W ++L +L +++ Q +ELGSG G+ G + +V+LTD
Sbjct: 183 LTGQLVWPGAMLLNDYLSKHIEM----FQGCTAIELGSGVGITGILCRRFCNKVVLTDHN 350
Query: 153 DR-LRLLRKNIETNM--KHVS-LRGSATATELTWGE-DPDRELIDPTP---DFVVGSDVV 204
+ L++++KNIE + +++S A +L WG D E++ P DFV+G+D+
Sbjct: 351 EEVLKIIKKNIELHSCPENISPTSNGLVAEKLEWGNTDQIHEILQKHPGGFDFVLGADIC 530
Query: 205 YSENAVVDLVETLGQL 220
+ ++ + L +T+ QL
Sbjct: 531 FQQSNIPLLFDTVRQL 578
>TC90766 similar to PIR|T05491|T05491 hypothetical protein T19K4.120 -
Arabidopsis thaliana, partial (63%)
Length = 1320
Score = 42.0 bits (97), Expect = 3e-04
Identities = 45/160 (28%), Positives = 74/160 (46%), Gaps = 7/160 (4%)
Frame = +1
Query: 94 TGAVM-WDSGVVLGKFLEHSVDLGTLVLQEKKIVELGSGCGLVGCIAALL--GGEVILTD 150
TG V W S VL + D + + KK++ELGSG GL G + A + EV+++D
Sbjct: 343 TGLVCNWPSEDVLAHYCLSHRD----IFRSKKVIELGSGYGLAGFVIAAITEASEVVISD 510
Query: 151 LLDR-LRLLRKNIETNMKHVSLRGSATATELTWGEDPDRELIDPTPDFVVGSDVVYSENA 209
+ + ++NIE N + +L W ++ + D D +V SD + ++
Sbjct: 511 GNPQVVDYTQRNIEANSGAFG-DTVVKSMKLHWNQEDTSSVADAF-DIIVASDCTFFKDF 684
Query: 210 VVDLVETLGQL---SGPNTTIFLAGELRNDAILEYFLEAA 246
DL + L + + IFL+ + N L+ FLE A
Sbjct: 685 HRDLARIVKHLLSKTESSEAIFLSPKRGNS--LDLFLEVA 798
>TC80790 similar to PIR|T38541|T38541 probable sucrose carrier - fission
yeast (Schizosaccharomyces pombe), partial (3%)
Length = 1461
Score = 36.6 bits (83), Expect = 0.011
Identities = 31/92 (33%), Positives = 46/92 (49%), Gaps = 2/92 (2%)
Frame = +3
Query: 92 GVTGAVMWDSGVVLGKFLEHSVDLGTLVLQEKKIVELGSGCGLVG-CIAALLGGEVILTD 150
G TG +W S + L + + +L K E+GSG GLVG C+A + + IL+D
Sbjct: 510 GDTGCSIWPSSLFLSELILSHPEL----FSNKVCFEIGSGVGLVGLCLAHVKASKGILSD 677
Query: 151 -LLDRLRLLRKNIETNMKHVSLRGSATATELT 181
L L ++ N+E N +V G+A E T
Sbjct: 678 GDLSTLANMKFNLELNNLNVE-TGTAQRNEDT 770
Score = 30.0 bits (66), Expect = 1.0
Identities = 13/51 (25%), Positives = 24/51 (46%)
Frame = +1
Query: 167 KHVSLRGSATATELTWGEDPDRELIDPTPDFVVGSDVVYSENAVVDLVETL 217
K + ++ L W + +L D PD ++G+DV+Y + LV +
Sbjct: 754 KEMKIQAPVKCMYLPWESASESQLQDIIPDVILGADVIYDPVCLPHLVRVI 906
>TC91651 SP|P40389|UV22_SCHPO UV-induced protein uvi22. [Fission yeast]
{Schizosaccharomyces pombe}, partial (3%)
Length = 655
Score = 34.7 bits (78), Expect = 0.041
Identities = 31/113 (27%), Positives = 53/113 (46%), Gaps = 2/113 (1%)
Frame = +2
Query: 72 LDSCGHSLSI-LQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLGTLVLQEKKIVELGS 130
+ SC +S + +Q + P V G +W + +VL F+ H L + +ELG+
Sbjct: 227 ISSCKNSYCVRIQHNITSSIPNV-GLQVWRAELVLTDFILHKA-LCSSEFHGVIALELGA 400
Query: 131 GCGLVGCIAALLGGEVILTDLLDR-LRLLRKNIETNMKHVSLRGSATATELTW 182
G GLVG + A V +TD ++ L KN++ N ++ + +L W
Sbjct: 401 GTGLVGLLLARTANSVFVTDRGNQILDNCVKNVQLNRGLLNNPATIFVRDLDW 559
>TC77387 similar to GP|15983396|gb|AAL11566.1 At1g51440/F5D21_19
{Arabidopsis thaliana}, partial (75%)
Length = 1793
Score = 32.3 bits (72), Expect = 0.20
Identities = 13/19 (68%), Positives = 17/19 (89%)
Frame = +3
Query: 2 ENRQEEEEEEEEEEDAPPP 20
E+ +EEEEEEEEEE++P P
Sbjct: 354 EDEEEEEEEEEEEEESPLP 410
Score = 31.2 bits (69), Expect = 0.46
Identities = 14/31 (45%), Positives = 19/31 (61%)
Frame = +3
Query: 2 ENRQEEEEEEEEEEDAPPPMVKLGSYGGEVR 32
E+ EEEEEEEEEE+ P+ L E++
Sbjct: 348 EDEDEEEEEEEEEEEEESPLPPLSEVWREIQ 440
>AL389387 similar to GP|9759518|dbj| gene_id:MLN1.9~pir||T38261~similar to
unknown protein {Arabidopsis thaliana}, partial (28%)
Length = 394
Score = 31.6 bits (70), Expect = 0.35
Identities = 21/68 (30%), Positives = 35/68 (50%), Gaps = 1/68 (1%)
Frame = +3
Query: 132 CGLVGCIAALLG-GEVILTDLLDRLRLLRKNIETNMKHVSLRGSATATELTWGEDPDREL 190
CG+ G LLG ++ILTD+ + L+KN+++N L+ + + L W
Sbjct: 3 CGVAGMGLYLLGLTDIILTDIPPVMPALKKNLKSNKP--VLKKNLKYSILYWNNKDQINA 176
Query: 191 IDPTPDFV 198
++P DFV
Sbjct: 177 VNPPFDFV 200
>TC84036 GP|493598|gb|AAA89204.1|| major immediate early protein {Gallid
herpesvirus 1}, partial (1%)
Length = 491
Score = 30.0 bits (66), Expect = 1.0
Identities = 13/13 (100%), Positives = 13/13 (100%)
Frame = -1
Query: 3 NRQEEEEEEEEEE 15
NRQEEEEEEEEEE
Sbjct: 365 NRQEEEEEEEEEE 327
>TC80247 similar to PIR|C86476|C86476 protein F15O4.45 [imported] -
Arabidopsis thaliana, partial (29%)
Length = 1225
Score = 30.0 bits (66), Expect = 1.0
Identities = 12/14 (85%), Positives = 13/14 (92%)
Frame = -2
Query: 8 EEEEEEEEDAPPPM 21
EE+EEEEED PPPM
Sbjct: 360 EEKEEEEEDFPPPM 319
>TC79991 similar to GP|10177307|dbj|BAB10568. contains similarity to zinc
finger protein~gene_id:MDC12.23 {Arabidopsis thaliana},
partial (8%)
Length = 1023
Score = 29.3 bits (64), Expect = 1.7
Identities = 12/14 (85%), Positives = 14/14 (99%)
Frame = -1
Query: 3 NRQEEEEEEEEEED 16
N+QEEEEEEEEEE+
Sbjct: 120 NQQEEEEEEEEEEE 79
>TC88708
Length = 478
Score = 29.3 bits (64), Expect = 1.7
Identities = 12/14 (85%), Positives = 14/14 (99%)
Frame = +2
Query: 3 NRQEEEEEEEEEED 16
NR+EEEEEEEEEE+
Sbjct: 35 NRREEEEEEEEEEE 76
Score = 27.3 bits (59), Expect = 6.6
Identities = 11/13 (84%), Positives = 13/13 (99%)
Frame = +2
Query: 4 RQEEEEEEEEEED 16
R+EEEEEEEEEE+
Sbjct: 41 REEEEEEEEEEEE 79
>TC83732 similar to GP|11762164|gb|AAG40360.1 AT4g22310 {Arabidopsis
thaliana}, partial (35%)
Length = 716
Score = 29.3 bits (64), Expect = 1.7
Identities = 12/14 (85%), Positives = 14/14 (99%)
Frame = +3
Query: 3 NRQEEEEEEEEEED 16
NR+EEEEEEEEEE+
Sbjct: 30 NRREEEEEEEEEEE 71
Score = 27.3 bits (59), Expect = 6.6
Identities = 11/13 (84%), Positives = 13/13 (99%)
Frame = +3
Query: 4 RQEEEEEEEEEED 16
R+EEEEEEEEEE+
Sbjct: 36 REEEEEEEEEEEE 74
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 28.9 bits (63), Expect = 2.3
Identities = 12/15 (80%), Positives = 13/15 (86%)
Frame = +3
Query: 2 ENRQEEEEEEEEEED 16
E +EEEEEEEEEED
Sbjct: 180 EEEEEEEEEEEEEED 224
Score = 27.7 bits (60), Expect = 5.0
Identities = 11/15 (73%), Positives = 13/15 (86%)
Frame = +3
Query: 2 ENRQEEEEEEEEEED 16
E +EEEEEEEEE+D
Sbjct: 183 EEEEEEEEEEEEEDD 227
Score = 27.3 bits (59), Expect = 6.6
Identities = 11/14 (78%), Positives = 13/14 (92%)
Frame = +3
Query: 3 NRQEEEEEEEEEED 16
+ +EEEEEEEEEED
Sbjct: 126 DEEEEEEEEEEEED 167
Score = 26.9 bits (58), Expect = 8.6
Identities = 11/15 (73%), Positives = 14/15 (93%)
Frame = +3
Query: 2 ENRQEEEEEEEEEED 16
E+ +EEEEEEEEEE+
Sbjct: 120 EHDEEEEEEEEEEEE 164
>TC83341 weakly similar to GP|8978076|dbj|BAA98104.1 gene_id:K14A3.4~unknown
protein {Arabidopsis thaliana}, partial (80%)
Length = 1253
Score = 28.9 bits (63), Expect = 2.3
Identities = 12/15 (80%), Positives = 13/15 (86%)
Frame = +1
Query: 2 ENRQEEEEEEEEEED 16
E +EEEEEEEEEED
Sbjct: 589 EEEEEEEEEEEEEED 633
Score = 27.7 bits (60), Expect = 5.0
Identities = 11/15 (73%), Positives = 13/15 (86%)
Frame = +1
Query: 2 ENRQEEEEEEEEEED 16
E +EEEEEEEEE+D
Sbjct: 592 EEEEEEEEEEEEEDD 636
Score = 27.7 bits (60), Expect = 5.0
Identities = 12/15 (80%), Positives = 13/15 (86%)
Frame = +1
Query: 2 ENRQEEEEEEEEEED 16
E QEEEEEEEEEE+
Sbjct: 574 EGLQEEEEEEEEEEE 618
Score = 27.3 bits (59), Expect = 6.6
Identities = 11/15 (73%), Positives = 13/15 (86%)
Frame = +1
Query: 2 ENRQEEEEEEEEEED 16
E +EEEEEEEEEE+
Sbjct: 586 EEEEEEEEEEEEEEE 630
Score = 26.9 bits (58), Expect = 8.6
Identities = 10/16 (62%), Positives = 14/16 (87%)
Frame = +1
Query: 1 MENRQEEEEEEEEEED 16
++ +EEEEEEEEEE+
Sbjct: 580 LQEEEEEEEEEEEEEE 627
>TC92853 similar to GP|20268748|gb|AAM14077.1 unknown protein {Arabidopsis
thaliana}, partial (21%)
Length = 761
Score = 28.9 bits (63), Expect = 2.3
Identities = 11/15 (73%), Positives = 14/15 (93%)
Frame = +3
Query: 2 ENRQEEEEEEEEEED 16
EN +EEE+E+EEEED
Sbjct: 384 ENEEEEEDEDEEEED 428
>CA860191 homologue to GP|19697342|gb hypothetical protein {Dictyostelium
discoideum}, partial (1%)
Length = 504
Score = 28.9 bits (63), Expect = 2.3
Identities = 11/16 (68%), Positives = 15/16 (93%)
Frame = +2
Query: 1 MENRQEEEEEEEEEED 16
++N +EEEEE+EEEED
Sbjct: 239 LQNDEEEEEEDEEEED 286
>TC91240 homologue to PIR|T48518|T48518 transcription factor like protein -
Arabidopsis thaliana, partial (7%)
Length = 680
Score = 28.5 bits (62), Expect = 3.0
Identities = 12/23 (52%), Positives = 17/23 (73%)
Frame = +3
Query: 1 MENRQEEEEEEEEEEDAPPPMVK 23
+E+ +EEEEEEEE+ D P +K
Sbjct: 267 VEDNEEEEEEEEEDADFNPLFLK 335
>BG447732 homologue to GP|9837385|gb|A retinitis pigmentosa GTPase
regulator-like protein {Takifugu rubripes}, partial (2%)
Length = 466
Score = 28.5 bits (62), Expect = 3.0
Identities = 11/15 (73%), Positives = 14/15 (93%)
Frame = -2
Query: 2 ENRQEEEEEEEEEED 16
EN +EEEEEEEE+E+
Sbjct: 120 ENEEEEEEEEEEDEE 76
>BQ136417 weakly similar to EGAD|146423|156 vitellogenin {Anolis pulchellus},
partial (20%)
Length = 621
Score = 28.1 bits (61), Expect = 3.9
Identities = 11/17 (64%), Positives = 14/17 (81%)
Frame = +2
Query: 2 ENRQEEEEEEEEEEDAP 18
E + EEEEEEEEE++P
Sbjct: 275 EEQSSEEEEEEEEEESP 325
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.135 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,742,402
Number of Sequences: 36976
Number of extensions: 99152
Number of successful extensions: 1536
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 656
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1144
length of query: 277
length of database: 9,014,727
effective HSP length: 95
effective length of query: 182
effective length of database: 5,502,007
effective search space: 1001365274
effective search space used: 1001365274
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0383.11


