

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0381.3
(253 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
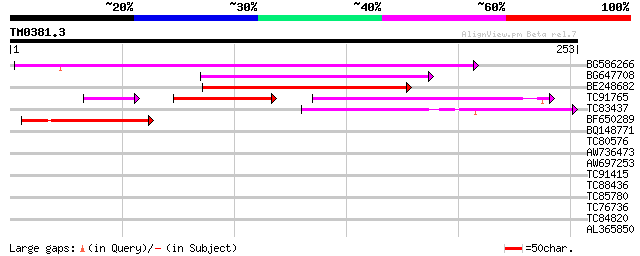
Score E
Sequences producing significant alignments: (bits) Value
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 122 1e-28
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 77 5e-15
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 69 1e-12
TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative ret... 43 1e-11
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 49 1e-06
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 45 2e-05
BQ148771 27 0.20
TC80576 similar to GP|11139266|gb|AAG31651.1 PRLI-interacting fa... 28 3.4
AW736473 similar to PIR|C84201|C842 multidrug resistance protein... 27 5.8
AW697253 similar to GP|12597289|gb| FasL isoform {Homo sapiens},... 27 5.8
TC91415 similar to GP|20856609|gb|AAM26675.1 At1g18620/F25I16_13... 27 5.8
TC88436 similar to GP|9293867|dbj|BAB01770.1 DEAD-Box RNA helica... 27 7.5
TC85780 similar to SP|P49608|ACOC_CUCMA Aconitate hydratase cyt... 27 7.5
TC76736 homologue to GP|22725683|gb|AAN04890.1 ribosomal protein... 27 9.9
TC84820 similar to GP|22136082|gb|AAM91119.1 GTP-binding protein... 27 9.9
AL365850 27 9.9
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 122 bits (306), Expect = 1e-28
Identities = 71/208 (34%), Positives = 109/208 (52%), Gaps = 1/208 (0%)
Frame = -3
Query: 3 ATRDNIILAQEVMHTIHTQ-KKGRGLVAIKIDLEKAYDRVSWDFLRATLYDFGFPPRIVD 61
A DN+++ +++H + K +A+K D+ KAYDR++W+FLR L GF +
Sbjct: 646 AISDNVLITHKILHYLRQSGAKKHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFHGIWIS 467
Query: 62 LIMWGVKCPKLSILWNCSKLPSFAPQRGLRQGDPLSPYLFVLCMERLAIQIQNLVTQGTW 121
IM V S L N P RGLRQGDPLSPYLF+LC E L+ Q + +GT
Sbjct: 466 WIMECVSTVSYSFLINGGPQGRVLPSRGLRQGDPLSPYLFILCTEVLSGLCQQALRKGTL 287
Query: 122 KPVRITKDGVGISHLFFADDVLLFCQASKEQLQLVNNTLKEFCEASGMKVNLDKSRMLFS 181
V++ ++ I+HL FADD + F +++ ++ + + ++ ASG +N KS + FS
Sbjct: 286 PGVKVARNCPPINHLLFADDTMFFGKSNASSCAILLSIMDKYRAASGRCIN*TKSAITFS 107
Query: 182 KAVDHRRQINLSHIAGISRASDLGKYLG 209
+ I++ GKYLG
Sbjct: 106 SKTSQAIIDRVKGELKIAKEGGTGKYLG 23
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 77.4 bits (189), Expect = 5e-15
Identities = 38/104 (36%), Positives = 62/104 (59%)
Frame = +1
Query: 86 PQRGLRQGDPLSPYLFVLCMERLAIQIQNLVTQGTWKPVRITKDGVGISHLFFADDVLLF 145
P++GLRQGDPLSPYLF+LC L+ ++ + +++ + I+HL FADD LLF
Sbjct: 4 PEKGLRQGDPLSPYLFILCANVLSGLLKREGNKQNLHGIQVARSDPKITHLLFADDSLLF 183
Query: 146 CQASKEQLQLVNNTLKEFCEASGMKVNLDKSRMLFSKAVDHRRQ 189
+A+ + + L + ASG VN +KS + +S+ V ++ +
Sbjct: 184 ARANLTEAATIMQVLHSYQSASGQLVNFEKSEVSYSQNVPNQEK 315
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 69.3 bits (168), Expect = 1e-12
Identities = 35/93 (37%), Positives = 57/93 (60%)
Frame = +3
Query: 87 QRGLRQGDPLSPYLFVLCMERLAIQIQNLVTQGTWKPVRITKDGVGISHLFFADDVLLFC 146
QRGL+QGDPL+P+LF+L E ++ ++N V + ++ + + G +SHL +ADD L
Sbjct: 21 QRGLKQGDPLAPFLFLLVAEGISGLMKNAVNRNLFQGFDVKRGGTRVSHLQYADDTLCIG 200
Query: 147 QASKEQLQLVNNTLKEFCEASGMKVNLDKSRML 179
+ + L + L+ F ASG+KVN KS ++
Sbjct: 201 MPTVDNLWTLKALLQGFEMASGLKVNFHKSSLI 299
>TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative retroelement
{Oryza sativa (japonica cultivar-group)}, partial (1%)
Length = 625
Score = 43.1 bits (100), Expect(3) = 1e-11
Identities = 19/46 (41%), Positives = 29/46 (62%)
Frame = +2
Query: 74 ILWNCSKLPSFAPQRGLRQGDPLSPYLFVLCMERLAIQIQNLVTQG 119
+L N + P RGL+QGD LSPY+F++C+E L+ I + +G
Sbjct: 128 VLVNNDAVDPIIPSRGLQQGDHLSPYIFIICVEGLSFLIPHAKERG 265
Score = 39.7 bits (91), Expect(3) = 1e-11
Identities = 31/109 (28%), Positives = 51/109 (46%), Gaps = 1/109 (0%)
Frame = +3
Query: 136 LFFADDVLLFCQASKEQLQLVNNTLKEFCEASGMKVNLDKSRMLFSKAVDHRRQINLSHI 195
L F L + + Q++ N L + E SG ++L KS + S+ V + ++++I
Sbjct: 279 LLFEGAPLRSLRVEEHHAQIMKNILILYEEDSGKAISLRKS*IYCSRNVPDILKTSITYI 458
Query: 196 AGISRASDLGKYLGIPLLKGRVTKAHFASLIGKMSSRLTSW-KNNLLNR 243
G+ KYLG+P + GR F+S+ G + W KN L R
Sbjct: 459 LGVQFMLGTCKYLGLPSMIGRDRTTTFSSIKGGV------WQKNKFLER 587
Score = 22.3 bits (46), Expect(3) = 1e-11
Identities = 9/25 (36%), Positives = 14/25 (56%)
Frame = +1
Query: 34 LEKAYDRVSWDFLRATLYDFGFPPR 58
+ K Y+RV D+L+ + GF R
Sbjct: 7 ISKVYNRVD*DYLKEIMIKMGFNNR 81
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 49.3 bits (116), Expect = 1e-06
Identities = 38/127 (29%), Positives = 62/127 (47%), Gaps = 4/127 (3%)
Frame = +2
Query: 131 VGISHLFFADDVLLFCQASKEQLQLVNNTLKEFCEASGMKVNLDKSRMLFSKAVDHRRQI 190
V +SHL FA+D LL + ++ + L F SG+KVN KS ++
Sbjct: 536 VVVSHLQFANDTLLLETKNWANIRALRAALVIF*AMSGLKVNFHKSGLVCVNIAPSW--- 706
Query: 191 NLSHIAGISRASDLGK----YLGIPLLKGRVTKAHFASLIGKMSSRLTSWKNNLLNRAGR 246
LS A + + +GK YLG+P+ + + ++ ++ +RLT W + L+ GR
Sbjct: 707 -LSEAASVL-SWKVGKVPFLYLGMPIEGNSRRLSFWEPIVNRIKARLTGWNSRFLSFGGR 880
Query: 247 VCLAKSV 253
+ L KSV
Sbjct: 881 LVLLKSV 901
Score = 33.1 bits (74), Expect = 0.11
Identities = 18/50 (36%), Positives = 26/50 (52%)
Frame = +1
Query: 6 DNIILAQEVMHTIHTQKKGRGLVAIKIDLEKAYDRVSWDFLRATLYDFGF 55
D I++A E + KK L+ K+D EKAY V W +L + L + F
Sbjct: 244 DGILIANEAVDEAKKLKKD--LLLFKVDFEKAYHSVDWAYLDSVLGRYVF 387
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 45.4 bits (106), Expect = 2e-05
Identities = 22/59 (37%), Positives = 37/59 (62%)
Frame = +3
Query: 6 DNIILAQEVMHTIHTQKKGRGLVAIKIDLEKAYDRVSWDFLRATLYDFGFPPRIVDLIM 64
DNIIL+ E++ + +++K +KIDL KAYD W F++ + + GFP + V+ +M
Sbjct: 384 DNIILSHELVKS-YSRKGISPRCMVKIDLXKAYDSXEWPFIKHLMLELGFPYKFVNWVM 557
>BQ148771
Length = 680
Score = 26.9 bits (58), Expect(2) = 0.20
Identities = 16/36 (44%), Positives = 21/36 (57%)
Frame = -3
Query: 218 TKAHFASLIGKMSSRLTSWKNNLLNRAGRVCLAKSV 253
TK S+ ++ L +WK N L+ A RV LAKSV
Sbjct: 618 TKKSRFSVYYQVHVMLANWKANHLSLARRVTLAKSV 511
Score = 23.9 bits (50), Expect(2) = 0.20
Identities = 9/16 (56%), Positives = 11/16 (68%)
Frame = -2
Query: 196 AGISRASDLGKYLGIP 211
AG A+ LGKY G+P
Sbjct: 679 AGFRXATSLGKYXGVP 632
>TC80576 similar to GP|11139266|gb|AAG31651.1 PRLI-interacting factor K
{Arabidopsis thaliana}, partial (41%)
Length = 933
Score = 28.1 bits (61), Expect = 3.4
Identities = 24/81 (29%), Positives = 40/81 (48%), Gaps = 2/81 (2%)
Frame = +3
Query: 25 RGLVAIKIDLEKAYDRVSWDFLRATLYDFGFPPRIVDLIMWGVKCPKLSILWNCSKLPS- 83
R L A+KI ++ +++ +F + L + V L+M+G+ C +LWN L S
Sbjct: 441 RDLQALKILTKRKWEQPIQEF*NSLLMKVQW----VFLLMYGIICFLKVVLWNLH*LKSA 608
Query: 84 -FAPQRGLRQGDPLSPYLFVL 103
F Q+GL Q L F++
Sbjct: 609 MFGFQKGLMQNFNLKEEDFLI 671
>AW736473 similar to PIR|C84201|C842 multidrug resistance protein homolog
[imported] - Halobacterium sp. NRC-1, partial (3%)
Length = 443
Score = 27.3 bits (59), Expect = 5.8
Identities = 9/16 (56%), Positives = 12/16 (74%)
Frame = -1
Query: 68 KCPKLSILWNCSKLPS 83
+CP S+ WNCS+ PS
Sbjct: 146 QCPSCSLDWNCSQCPS 99
>AW697253 similar to GP|12597289|gb| FasL isoform {Homo sapiens}, partial
(16%)
Length = 761
Score = 27.3 bits (59), Expect = 5.8
Identities = 11/28 (39%), Positives = 16/28 (56%)
Frame = -1
Query: 69 CPKLSILWNCSKLPSFAPQRGLRQGDPL 96
CP+ S S P+F PQR ++ G P+
Sbjct: 530 CPRASSAPTTSPAPTFPPQRHIKHGFPI 447
>TC91415 similar to GP|20856609|gb|AAM26675.1 At1g18620/F25I16_13
{Arabidopsis thaliana}, partial (3%)
Length = 705
Score = 27.3 bits (59), Expect = 5.8
Identities = 16/50 (32%), Positives = 26/50 (52%)
Frame = +2
Query: 143 LLFCQASKEQLQLVNNTLKEFCEASGMKVNLDKSRMLFSKAVDHRRQINL 192
+L Q S+EQL ++N + FC+ G +V+L + F +D NL
Sbjct: 128 ILNSQHSEEQLSEISNDSRSFCQ--GDEVSLQSDSVTFDSKLDIEVTSNL 271
>TC88436 similar to GP|9293867|dbj|BAB01770.1 DEAD-Box RNA helicase-like
protein {Arabidopsis thaliana}, partial (38%)
Length = 1024
Score = 26.9 bits (58), Expect = 7.5
Identities = 11/19 (57%), Positives = 14/19 (72%)
Frame = +2
Query: 72 LSILWNCSKLPSFAPQRGL 90
LS+LW + PSF+P RGL
Sbjct: 338 LSLLWRRKESPSFSPFRGL 394
>TC85780 similar to SP|P49608|ACOC_CUCMA Aconitate hydratase cytoplasmic (EC
4.2.1.3) (Citrate hydro-lyase) (Aconitase)., partial
(97%)
Length = 3124
Score = 26.9 bits (58), Expect = 7.5
Identities = 15/43 (34%), Positives = 21/43 (47%)
Frame = -1
Query: 67 VKCPKLSILWNCSKLPSFAPQRGLRQGDPLSPYLFVLCMERLA 109
V CP L L+ SK +F+P+ P SP+ + M LA
Sbjct: 2389 VSCPSLLYLFAASKTDNFSPEGTCIVLGPTSPFRNLFTMRILA 2261
>TC76736 homologue to GP|22725683|gb|AAN04890.1 ribosomal protein L14 {Vigna
angularis}, partial (38%)
Length = 566
Score = 26.6 bits (57), Expect = 9.9
Identities = 14/47 (29%), Positives = 22/47 (46%)
Frame = -1
Query: 33 DLEKAYDRVSWDFLRATLYDFGFPPRIVDLIMWGVKCPKLSILWNCS 79
+L+K + R+ W Y G R VDL+ K ++W+CS
Sbjct: 401 NLKKRFPRLGWPKSFGIRYHGG*ESRSVDLMTLYDTLSKSRVVWDCS 261
>TC84820 similar to GP|22136082|gb|AAM91119.1 GTP-binding protein LepA-like
protein {Arabidopsis thaliana}, partial (32%)
Length = 926
Score = 26.6 bits (57), Expect = 9.9
Identities = 15/45 (33%), Positives = 25/45 (55%), Gaps = 1/45 (2%)
Frame = +3
Query: 103 LCMERLAIQIQNLVTQGTWKPVRITKD-GVGISHLFFADDVLLFC 146
+CM A+ NL+ + +K + T D + +SHLFF+ +L C
Sbjct: 675 VCM*VAALFGCNLINKHEYKRMIYTSDESLIVSHLFFSSPYILLC 809
>AL365850
Length = 568
Score = 26.6 bits (57), Expect = 9.9
Identities = 12/25 (48%), Positives = 14/25 (56%)
Frame = +1
Query: 66 GVKCPKLSILWNCSKLPSFAPQRGL 90
GVKCP + W LP FAP G+
Sbjct: 94 GVKCPLVQFSW----LPLFAPDMGI 156
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.139 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,229,918
Number of Sequences: 36976
Number of extensions: 114917
Number of successful extensions: 655
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 651
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 653
length of query: 253
length of database: 9,014,727
effective HSP length: 94
effective length of query: 159
effective length of database: 5,538,983
effective search space: 880698297
effective search space used: 880698297
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0381.3


