

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0378b.9
(768 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
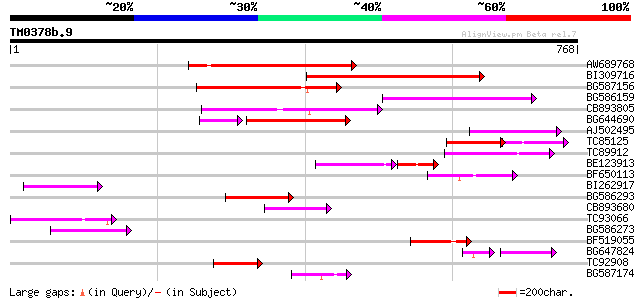
Score E
Sequences producing significant alignments: (bits) Value
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 252 3e-67
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 187 1e-47
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 164 1e-40
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 161 8e-40
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 156 2e-38
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 129 1e-37
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 106 4e-23
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 67 2e-21
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 98 1e-20
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 77 4e-20
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 92 1e-18
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 80 4e-15
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 74 2e-13
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 73 5e-13
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 71 1e-12
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 70 3e-12
BF519055 similar to GP|21592754|gb| unknown {Arabidopsis thalian... 62 6e-10
BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F... 55 3e-09
TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol po... 59 7e-09
BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse ... 59 9e-09
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 252 bits (644), Expect = 3e-67
Identities = 123/227 (54%), Positives = 156/227 (68%)
Frame = +1
Query: 243 TRAKTGIHRPKAYAASTALSSVPTSAK*ALSIPHWHQAMQEEYQALMNNNTWELVPLLHG 302
TR KTG +PK + LS P W QAM+ EY+AL++N TW+LVPL
Sbjct: 4 TRTKTGKLKPKVFLTEPCPQDCENLP---LSDPRWLQAMKTEYKALIDNKTWDLVPLPPH 174
Query: 303 RDAIGCKWVFRTKYNSDGSINKHKPRLVAKGFHQTEGYDYNETFSPVVKPTTIRTVLSLA 362
+ AIGCKWV+R K N DGS+NK K RLVAKGF QT G DY ETFSPV+KP TIR +L++A
Sbjct: 175 KKAIGCKWVYRVKENPDGSVNKFKARLVAKGFSQTLGCDYTETFSPVIKPVTIRLILTIA 354
Query: 363 LMHQWPIRQFDFNNAFLNGELVEEVFMQQPPGFSKGSSHLVCKLKKALYGLKQAPRAWFD 422
+ ++W I+Q D NNAFLNG L EEV+M QP GF + LVCKL K+LYGLKQAPRAW++
Sbjct: 355 ITYKWEIQQIDINNAFLNGFLQEEVYMSQPQGFEAANKSLVCKLNKSLYGLKQAPRAWYE 534
Query: 423 KLSSTLARFGFQAAKCDTSLFCKFTKSETLYILVYVDDIIITGSSST 469
L+S +FGF ++CD SL +Y+ +YVDDI+ITGSS++
Sbjct: 535 XLTSAQIQFGFTKSRCDPSLLIYNQNGACIYLXIYVDDILITGSSAS 675
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 187 bits (475), Expect = 1e-47
Identities = 96/241 (39%), Positives = 148/241 (60%)
Frame = +2
Query: 403 VCKLKKALYGLKQAPRAWFDKLSSTLARFGFQAAKCDTSLFCKFTKSETLYILVYVDDII 462
VC+L+K++YGLKQA R W+ KLS +L FG+ + D SLF KF S +LVYVDDI+
Sbjct: 20 VCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDIV 199
Query: 463 ITGSSSTAIFSLIADLNAVFPLKDLGSLHYFLGIEATHTKDGGLILSQTKYINDLLIKAK 522
+ G+ + I + L F +KDLGSL YFLG+E +K G ++L+Q KY +LL +
Sbjct: 200 LAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQG-ILLNQRKYTLELLEDSG 376
Query: 523 MDSINPSPTPMTSGLKLSANSGALFPKPLLYRSVVGALHYVTITRPEIAFPLNKVCQFMH 582
++ + TP LKL + L+ YR ++G L Y+T TRP+I+F + ++ QF+
Sbjct: 377 NLAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFVS 556
Query: 583 SPLEPHWQAVKRILRYLHGTATHGLYLRRPSSMSITAFADSDWGSDPDDRRSTSGMCIYI 642
P + H+QA R+L+YL GL+ S++ +++FADSDW + P R+S +G +++
Sbjct: 557 KPQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGYWVFL 736
Query: 643 G 643
G
Sbjct: 737 G 739
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 164 bits (415), Expect = 1e-40
Identities = 90/201 (44%), Positives = 124/201 (60%), Gaps = 5/201 (2%)
Frame = -1
Query: 254 AYAASTALSSVPTSAK*ALSIPHWHQAMQEEYQALMNNNTWELVPLLHGRDAIGCKWVFR 313
A+ + + +P S + A+ W +++ E A++ N+TW L G+ A+ +W+F
Sbjct: 591 AFMVNLNENHIPRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSSRWIFT 412
Query: 314 TKYNSDGSINKHKPRLVAKGFHQTEGYDYNETFSPVVKPTTIRTVLSLALMHQWPIRQFD 373
KY +DGSI + K RLVA+GF T G DY ETF+PV K TIR VLSLA+ W + Q D
Sbjct: 411 IKYKADGSIERKKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQMD 232
Query: 374 FNNAFLNGELVEEVFMQQPPGFSKGSSHL-----VCKLKKALYGLKQAPRAWFDKLSSTL 428
NAFL GEL +EV+M PPG HL V +LKKA+YGLKQ+PRAW++KLS+TL
Sbjct: 231 VKNAFLQGELEDEVYMYPPPGL----EHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTL 64
Query: 429 ARFGFQAAKCDTSLFCKFTKS 449
GF+ ++ D +LF T S
Sbjct: 63 NGRGFRKSELDHTLFTLTTPS 1
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 161 bits (408), Expect = 8e-40
Identities = 81/209 (38%), Positives = 122/209 (57%)
Frame = +1
Query: 505 GLILSQTKYINDLLIKAKMDSINPSPTPMTSGLKLSANSGALFPKPLLYRSVVGALHYVT 564
G+ + Q KY+ DLL + M+ N S P+ KL + + Y+ +VG L Y+
Sbjct: 25 GIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQIVGCLMYLA 204
Query: 565 ITRPEIAFPLNKVCQFMHSPLEPHWQAVKRILRYLHGTATHGLYLRRPSSMSITAFADSD 624
TRP++ + L+ + +FM+ P E H AVKR+LRYL+GT G+ +R S + A+ DSD
Sbjct: 205 ATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEKLEAYTDSD 384
Query: 625 WGSDPDDRRSTSGMCIYIGGNLVSWAAKKQTVVARSSTEAEYRSLALVITEIMWLRSLLS 684
+ D DDR+STSG + VSW++KKQ VV S+T+AE+ + A + +W+R +L
Sbjct: 385 YAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQSVWMRRVLE 564
Query: 685 ELRVPTSPSSLIYCDNLSVVHLVANPILH 713
+L S S +YCDN S + L NP+LH
Sbjct: 565 KLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 156 bits (395), Expect = 2e-38
Identities = 90/252 (35%), Positives = 133/252 (52%), Gaps = 8/252 (3%)
Frame = +3
Query: 261 LSSVPTSAK*ALSIPHWHQAMQEEYQALMNNNTWELVPLLHGRDAIGCKWVFRTKYNSDG 320
++S PT+ + A+ W +M E +A NNTWEL L G IG KW+F+TK N +G
Sbjct: 30 MTSDPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNENG 209
Query: 321 SINKHKPRLVAKGFHQTEGYDYNETFSPVVKPTTIRTVLSLALMHQWPIRQFDFNNAFLN 380
I K+K RLVAKG+ Q G DY E F+PV + TIR V++LA Q + ++
Sbjct: 210 EIEKYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALA-------AQIKRDGVCIS 368
Query: 381 GELVEEVFMQQPPGFSKGSSHLVC-------KLKKALYGLKQAPRAWFDKLSSTLARFGF 433
M+ +H V ++K+ALYGLKQAPRAW+ ++ + + GF
Sbjct: 369 *M*KAHSCMEN*MRKFLLINHRVM*RRVIS*RVKRALYGLKQAPRAWYSRIEAYFTKEGF 548
Query: 434 QAAKCDTSLFCKFTK-SETLYILVYVDDIIITGSSSTAIFSLIADLNAVFPLKDLGSLHY 492
+ + +LF K ++ + L I +YVDD+I G+ + F + DLG +HY
Sbjct: 549 EKCPYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGKMHY 728
Query: 493 FLGIEATHTKDG 504
FLG+E T + G
Sbjct: 729 FLGVEVTQNEKG 764
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 129 bits (325), Expect(2) = 1e-37
Identities = 66/141 (46%), Positives = 93/141 (65%), Gaps = 1/141 (0%)
Frame = -2
Query: 322 INKHKPRLVAKGFHQTEGYDYNETFSPVVKPTTIRTVLSLALMHQWPIRQFDFNNAFLNG 381
I ++K +LV +G++Q EG DY+E FSPV + IR +++ A + + Q D +AF+NG
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 382 ELVEEVFMQQPPGFSKGS-SHLVCKLKKALYGLKQAPRAWFDKLSSTLARFGFQAAKCDT 440
+L EEVF++QPPGF + V +L K LYGLKQAPRAW+++LS L + GF+ K D
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 441 SLFCKFTKSETLYILVYVDDI 461
+LF + E L I VYVDDI
Sbjct: 64 TLFLLKRE*ELLIIQVYVDDI 2
Score = 45.8 bits (107), Expect(2) = 1e-37
Identities = 24/59 (40%), Positives = 31/59 (51%), Gaps = 1/59 (1%)
Frame = -3
Query: 258 STALSSV-PTSAK*ALSIPHWHQAMQEEYQALMNNNTWELVPLLHGRDAIGCKWVFRTK 315
S +SS+ P + K AL W +MQEE + W LVP G+ IG +WVFR K
Sbjct: 609 SAFISSIEPKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKTVIGTRWVFRNK 433
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 106 bits (264), Expect = 4e-23
Identities = 47/125 (37%), Positives = 75/125 (59%)
Frame = +2
Query: 623 SDWGSDPDDRRSTSGMCIYIGGNLVSWAAKKQTVVARSSTEAEYRSLALVITEIMWLRSL 682
SDW D + R+STSG ++G +SW++KKQ VVA S+ EAEY + T+ +WLR +
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 683 LSELRVPTSPSSLIYCDNLSVVHLVANPILHTKTKHFALDLHFVRERVADKQVTVVNIPA 742
L + + + IYCDN S + L NP+ H ++KH + H +RE +A+K+V + P
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 743 PSQVS 747
+++
Sbjct: 362 EEKIA 376
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 66.6 bits (161), Expect(2) = 2e-21
Identities = 35/80 (43%), Positives = 50/80 (61%)
Frame = +1
Query: 592 VKRILRYLHGTATHGLYLRRPSSMSITAFADSDWGSDPDDRRSTSGMCIYIGGNLVSWAA 651
VKRI+RY+ GT+ + S +++ + DSD+ D D R+ST+G + G VSW +
Sbjct: 1 VKRIMRYIKGTSGVAVCFGG-SELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLS 177
Query: 652 KKQTVVARSSTEAEYRSLAL 671
K QTVVA S+TEAEY + L
Sbjct: 178 KLQTVVALSTTEAEYMAAYL 237
Score = 54.7 bits (130), Expect(2) = 2e-21
Identities = 27/89 (30%), Positives = 44/89 (49%)
Frame = +3
Query: 668 SLALVITEIMWLRSLLSELRVPTSPSSLIYCDNLSVVHLVANPILHTKTKHFALDLHFVR 727
SL E +W++ L+ EL + +YCD+ S +H+ NP H++TKH + HFVR
Sbjct: 228 SLPQACKEAIWMQRLMEELGHKQEQIT-VYCDSQSALHIARNPAFHSRTKHIGIQYHFVR 404
Query: 728 ERVADKQVTVVNIPAPSQVSSTLFHCFKT 756
E V + V + I ++ + T
Sbjct: 405 EVVEEGSVDMQKIHTNDNLADAMTKSINT 491
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 98.2 bits (243), Expect = 1e-20
Identities = 47/150 (31%), Positives = 87/150 (57%), Gaps = 2/150 (1%)
Frame = +1
Query: 590 QAVKRILRYLHGTATHGLYLRRPSSM--SITAFADSDWGSDPDDRRSTSGMCIYIGGNLV 647
QA+K +L+YL+ + L + + ++ + D+D+ + D R+S SG + G +
Sbjct: 1 QALKWVLKYLNESLKSSLKYTKAAQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTTI 180
Query: 648 SWAAKKQTVVARSSTEAEYRSLALVITEIMWLRSLLSELRVPTSPSSLIYCDNLSVVHLV 707
SW A +Q+VV S+T+AEY + + + +WL+ ++ EL + T I+CD+ S +HL
Sbjct: 181 SWKANQQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGI-TQEYVKIHCDSQSAIHLA 357
Query: 708 ANPILHTKTKHFALDLHFVRERVADKQVTV 737
+ + H +TKH + LHF+R+ + K++ V
Sbjct: 358 NHQVYHERTKHIDIRLHFIRDMIESKEIVV 447
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 77.4 bits (189), Expect(2) = 4e-20
Identities = 44/110 (40%), Positives = 65/110 (59%), Gaps = 1/110 (0%)
Frame = +1
Query: 415 QAPRAWFDKLSSTLARFGFQAAKCDTSLFCKFTKS-ETLYILVYVDDIIITGSSSTAIFS 473
Q+PR WFD+ + + +FG+ + D ++F K + + + ++VYVDDI +TG I
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 474 LIADLNAVFPLKDLGSLHYFLGIEATHTKDGGLILSQTKYINDLLIKAKM 523
L L F +KDLG+L YFLG+E K G I SQ KY+ DLL + +M
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSI-SQRKYVLDLLKETRM 327
Score = 39.3 bits (90), Expect(2) = 4e-20
Identities = 23/57 (40%), Positives = 36/57 (62%), Gaps = 1/57 (1%)
Frame = +2
Query: 526 INPSPTPMTSGLKLSA-NSGALFPKPLLYRSVVGALHYVTITRPEIAFPLNKVCQFM 581
+ PS TPM + +KL ++G L K Y+ +VG L Y++ TRP+I+F + + QFM
Sbjct: 335 VKPSETPMDATVKLGTLDNGTLVDKGR-YQRLVGKLIYLSHTRPDISFVVCTMSQFM 502
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 91.7 bits (226), Expect = 1e-18
Identities = 48/124 (38%), Positives = 69/124 (54%), Gaps = 3/124 (2%)
Frame = +1
Query: 567 RPEIAFPLNKVCQFMHSPLEPHWQAVKRILRYLHGTATHGL---YLRRPSSMSITAFADS 623
RP+I + ++ + +FMH P +PH A RILRY+ GT +GL Y + + ++DS
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICYSDS 300
Query: 624 DWGSDPDDRRSTSGMCIYIGGNLVSWAAKKQTVVARSSTEAEYRSLALVITEIMWLRSLL 683
DW D RRSTSG +SW KKQ + A SS EAEY + + +WL S++
Sbjct: 301 DWCGD---RRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSVI 471
Query: 684 SELR 687
EL+
Sbjct: 472 KELK 483
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 79.7 bits (195), Expect = 4e-15
Identities = 44/107 (41%), Positives = 57/107 (53%)
Frame = +1
Query: 19 QQNGRVEWKHRHITETGLALLAHAHMPLTFWGDAFETAAYLINQLPIPTLDLSTPLQKLY 78
QQNG E +R + E A+L A M +FW +A +TA Y+IN+ P +DL TP++
Sbjct: 91 QQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACYVINRSPSTVIDLKTPMEMWK 270
Query: 79 DKKPDYSFLKCFGCLCYPHTRPYNTHKLAFRSAPCTFLGYSPMHKGY 125
K DYS L FGC Y KL +S C FLGY+ KGY
Sbjct: 271 GKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYADNVKGY 411
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 74.3 bits (181), Expect = 2e-13
Identities = 39/92 (42%), Positives = 57/92 (61%)
Frame = +2
Query: 293 TWELVPLLHGRDAIGCKWVFRTKYNSDGSINKHKPRLVAKGFHQTEGYDYNETFSPVVKP 352
T +LV G IG +W+++ K N DG++ K+K RLVAKG+ + +G D++E F+PVV+
Sbjct: 56 TLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPVVRI 235
Query: 353 TTIRTVLSLALMHQWPIRQFDFNNAFLNGELV 384
TI +L+LA + I D AFLNG V
Sbjct: 236 ETI*LLLALAATNGC*IHHIDVKIAFLNGHFV 331
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 72.8 bits (177), Expect = 5e-13
Identities = 38/90 (42%), Positives = 53/90 (58%)
Frame = -2
Query: 346 FSPVVKPTTIRTVLSLALMHQWPIRQFDFNNAFLNGELVEEVFMQQPPGFSKGSSHLVCK 405
F P+VK TI +LS+ + + D AFL G+LVE+++M QP GFS +V K
Sbjct: 554 FVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGFS*EVGKMVGK 375
Query: 406 LKKALYGLKQAPRAWFDKLSSTLARFGFQA 435
LKK++YGLKQ PR L + R GF++
Sbjct: 374 LKKSMYGLKQGPRQCI*SLKALCTRKGFRS 285
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 71.2 bits (173), Expect = 1e-12
Identities = 51/148 (34%), Positives = 70/148 (46%), Gaps = 5/148 (3%)
Frame = +1
Query: 2 FLTQCGIQHRLTCPYVHQQNGRVEWKHRHITETGLALLAHAHMP--LTFWGDAFETAAYL 59
F T GI T P QQNG E R + E +L++A + W +A TA +L
Sbjct: 343 FCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHL 522
Query: 60 INQLPIPTLDLSTPLQKLYDKKPDYSFLKCFGCLCYPHTRPYNTHKLAFRSAPCTFLGYS 119
+N+ P LD P DYS L+ FGC Y N KLA R+ C FL Y+
Sbjct: 523 VNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYALV---NDGKLAPRAGECIFLSYA 693
Query: 120 PMHKGYNCLTTE---GKVIITRNVLFDE 144
KGY ++ K+I++R+V F+E
Sbjct: 694 SESKGYRLWCSDPKSQKLILSRDVTFNE 777
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 70.1 bits (170), Expect = 3e-12
Identities = 37/111 (33%), Positives = 58/111 (51%), Gaps = 1/111 (0%)
Frame = -2
Query: 56 AAYLINQLPIPTLDLSTPLQKLYDKKPDYSFLKCFGCLCYPHTRPYNTHKLAFRSAPCTF 115
A YLIN++P L P + L +KP ++++ FGCLCY +KL RS F
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMF 525
Query: 116 LGYSPMHKGYNCLTTEG-KVIITRNVLFDETVFPFQFQTSKSSSDILLDSS 165
+GYS KGY C E +V+++R+V F E ++ + + D+ D +
Sbjct: 524 IGYSTTQKGYKCYDPEARRVLVSRDVKFIEERGYYEEKNQEDLRDLTSDKA 372
>BF519055 similar to GP|21592754|gb| unknown {Arabidopsis thaliana}, partial
(30%)
Length = 675
Score = 62.4 bits (150), Expect = 6e-10
Identities = 29/82 (35%), Positives = 52/82 (63%)
Frame = -3
Query: 544 GALFPKPLLYRSVVGALHYVTITRPEIAFPLNKVCQFMHSPLEPHWQAVKRILRYLHGTA 603
G++ L S+VGAL Y+T+T P+++F +NK QFMH P + ++Q +K+++R+
Sbjct: 661 GSITADSKLVHSIVGALLYITVTCPDLSFSINKPSQFMHKPTQINFQQLKKVMRH----- 497
Query: 604 THGLYLRRPSSMSITAFADSDW 625
L +++ + I AF+D+DW
Sbjct: 496 -PKLTIKKLFDLQIYAFSDADW 434
>BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (5%)
Length = 721
Score = 54.7 bits (130), Expect(2) = 3e-09
Identities = 29/76 (38%), Positives = 43/76 (56%), Gaps = 1/76 (1%)
Frame = -3
Query: 666 YRSLALVITEIMWLRSLLSELRVPTSPSSLIYCDNLSVV-HLVANPILHTKTKHFALDLH 724
YRS+ I EI WL LL++L+ +++YCDN S H+ AN +TKH LD H
Sbjct: 374 YRSI*STICEIKWLTYLLNDLKFTFIKPAMLYCDNQSAARHIAANSSFLERTKHIELDCH 195
Query: 725 FVRERVADKQVTVVNI 740
VR ++ K +++I
Sbjct: 194 IVRVKLQLKLFHILHI 147
Score = 25.4 bits (54), Expect(2) = 3e-09
Identities = 16/52 (30%), Positives = 26/52 (49%), Gaps = 9/52 (17%)
Frame = -2
Query: 614 SMSITAFADSDWG---------SDPDDRRSTSGMCIYIGGNLVSWAAKKQTV 656
+M+I + DSD S D +S S CI++G +L+ W + K+ V
Sbjct: 543 TMNIFSILDSDQSIL*LRLD*SSCLDT*KSISYFCIFLGDSLICWKS*KKKV 388
>TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol polyprotein
{Oryza sativa}, partial (1%)
Length = 638
Score = 58.9 bits (141), Expect = 7e-09
Identities = 28/66 (42%), Positives = 41/66 (61%)
Frame = +2
Query: 277 WHQAMQEEYQALMNNNTWELVPLLHGRDAIGCKWVFRTKYNSDGSINKHKPRLVAKGFHQ 336
W AM++E Q L NNT L PL G+ + C+ V++ + ++G I K+K +LVAK F Q
Sbjct: 317 WLAAMEQEIQVLEENNTSTLEPLREGKKWVDCRPVYKIIHKANGEIEKYKAQLVAKDFVQ 496
Query: 337 TEGYDY 342
EG D+
Sbjct: 497 VEGEDF 514
>BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse
transcriptase homolog - rape retrotransposon copia-like
(fragment), partial (84%)
Length = 249
Score = 58.5 bits (140), Expect = 9e-09
Identities = 35/85 (41%), Positives = 49/85 (57%), Gaps = 4/85 (4%)
Frame = -1
Query: 382 ELVEEVFMQQPPGFS-KGSSHLVCKLKKALYGLKQAPRAW---FDKLSSTLARFGFQAAK 437
EL E+++M QP GF G VCKL+K+LYGLKQ+PR W FD S+ A G
Sbjct: 249 ELEEKIYMTQPEGFLFPGKEDHVCKLRKSLYGLKQSPRQWYKRFDSYRSSWATTGVLMTV 70
Query: 438 CDTSLFCKFTKSETLYILVYVDDII 462
T + S +Y+++YVDD++
Sbjct: 69 VST--*TR*RMSRYIYLVLYVDDML 1
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.135 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,843,523
Number of Sequences: 36976
Number of extensions: 436142
Number of successful extensions: 2567
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 2515
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2552
length of query: 768
length of database: 9,014,727
effective HSP length: 104
effective length of query: 664
effective length of database: 5,169,223
effective search space: 3432364072
effective search space used: 3432364072
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0378b.9


