

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0378b.5
(572 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
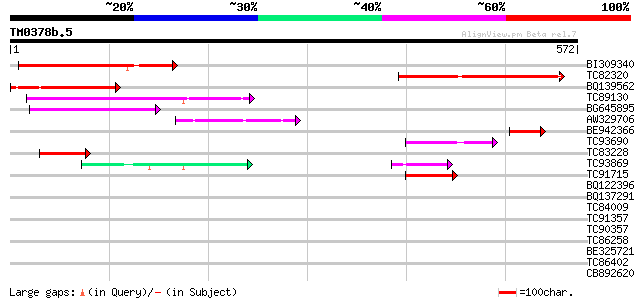
Score E
Sequences producing significant alignments: (bits) Value
BI309340 weakly similar to GP|13605829|gb AT5g04550/T32M21_140 {... 146 2e-35
TC82320 similar to GP|8953735|dbj|BAA98099.1 contains similarity... 143 2e-34
BQ139562 128 5e-30
TC89130 similar to GP|12003396|gb|AAG43555.1 Avr9/Cf-9 rapidly e... 107 1e-23
BG645895 weakly similar to GP|7939524|dbj contains similarity to... 96 4e-20
AW329706 weakly similar to GP|7939524|dbj| contains similarity t... 54 2e-07
BE942366 similar to GP|7939524|dbj| contains similarity to unkno... 52 5e-07
TC93690 similar to PIR|D86467|D86467 protein F23M19.3 [imported]... 52 8e-07
TC83228 weakly similar to GP|8953735|dbj|BAA98099.1 contains sim... 50 2e-06
TC93869 weakly similar to GP|20466802|gb|AAM20718.1 unknown prot... 50 2e-06
TC91715 weakly similar to GP|20466802|gb|AAM20718.1 unknown prot... 47 2e-05
BQ122396 similar to GP|7939524|dbj| contains similarity to unkno... 39 0.007
BQ137291 similar to GP|13605829|gb| AT5g04550/T32M21_140 {Arabid... 33 0.30
TC84009 similar to GP|13548324|emb|CAC35871. putative protein {A... 33 0.39
TC91357 similar to GP|2194137|gb|AAB61112.1| ESTs gb|R29947 gb|H... 32 0.66
TC90357 similar to GP|20466802|gb|AAM20718.1 unknown protein {Ar... 32 0.66
TC86258 similar to GP|16930433|gb|AAL31902.1 AT4g17230/dl4650c {... 32 0.66
BE325721 similar to SP|P49147|PA2Y_ Cytosolic phospholipase A2 (... 30 2.5
TC86402 homologue to GP|15148926|gb|AAK84890.1 TGA-type basic le... 29 4.3
CB892620 weakly similar to GP|22655012|g At1g73080/F3N23_28 {Ara... 28 7.3
>BI309340 weakly similar to GP|13605829|gb AT5g04550/T32M21_140 {Arabidopsis
thaliana}, partial (27%)
Length = 574
Score = 146 bits (368), Expect = 2e-35
Identities = 72/164 (43%), Positives = 108/164 (64%), Gaps = 4/164 (2%)
Frame = +2
Query: 10 SWLSALWPVSRKGASDNKAVVVGVLGSEVAGLMLKVVNLWQSLSDEEVLSLREGIVNSAG 69
SW +LW RK ++++ V+GVL E+A LM K+VNLWQSLSD+ + L+E I NS G
Sbjct: 95 SWFRSLWRTPRKHDANSEKEVIGVLAFEIASLMSKLVNLWQSLSDKHISRLKEEITNSIG 274
Query: 70 VRKLVSEDDDYLMELVLNEVLDNFQSVARTVARLGRRCVDPVYHRFEH----FVRSPAQN 125
++KLVS+DD ++ L+ E+++N VA +VARL ++C DP+ FE+ F+ +
Sbjct: 275 IKKLVSDDDHFIERLICMEIVENMAHVAESVARLAKKCNDPILKGFENTFYGFITTGTDP 454
Query: 126 YFQWSGWDYRGKKMERKVKRMEKFVAAMTQLCQELEVLAEVEQT 169
Y GW+ KKME+K+K+ EKF++ L QE+EVL ++EQT
Sbjct: 455 Y----GWELTCKKMEKKIKKFEKFISTNASLYQEMEVLVDLEQT 574
>TC82320 similar to GP|8953735|dbj|BAA98099.1 contains similarity to unknown
protein~emb|CAB85560.1~gene_id:K10D11.2 {Arabidopsis
thaliana}, partial (30%)
Length = 875
Score = 143 bits (361), Expect = 2e-34
Identities = 80/168 (47%), Positives = 110/168 (64%), Gaps = 1/168 (0%)
Frame = +1
Query: 393 LKPASFTLGDAALTLHYANMIVLIEQMVSSPHLTDPEPRDDLYKMLPATIRAVLRHKLKG 452
LKP +LGD+ L+LHYAN+I+++E+M+ SP L + RDDLY MLP +IR+ LR +LKG
Sbjct: 187 LKPPKGSLGDSGLSLHYANLIIVMEKMIKSPQLVGVDARDDLYAMLPNSIRSSLRLRLKG 366
Query: 453 RKKSKSSWVYNAEIAAEWSAVLAQILDWLAPLAHDTIRWHSVRSF-EKEHATLKANILLV 511
S + +A EW L +IL WL PLA + IRW S RS EK K+N+LL+
Sbjct: 367 ---SIGFSACDPLLANEWKNALGRILCWLLPLAQNMIRWQSERSVEEKSLVPKKSNVLLL 537
Query: 512 QTLYFANQVKTEAAIVDLLVGLNYVCRIDTKVGVRDRVECASARSFHG 559
QTL FA++ KTEAAI +LLVGLNYV R + ++ + +C +F+G
Sbjct: 538 QTLVFADKAKTEAAITELLVGLNYVWRFEREMTAKALFQCTD--NFNG 675
>BQ139562
Length = 389
Score = 128 bits (322), Expect = 5e-30
Identities = 64/111 (57%), Positives = 86/111 (76%)
Frame = +3
Query: 1 MGGETVNGSSWLSALWPVSRKGASDNKAVVVGVLGSEVAGLMLKVVNLWQSLSDEEVLSL 60
M GETVN +WL +WPVSRK SD + G++ EVAGLM KVVNLW SLSD E+++L
Sbjct: 63 MKGETVN-VTWLGGIWPVSRKSGSDENNEI-GIMAFEVAGLMSKVVNLWHSLSDNELMNL 236
Query: 61 REGIVNSAGVRKLVSEDDDYLMELVLNEVLDNFQSVARTVARLGRRCVDPV 111
RE IV+S GV+ LVS+D+ +LMEL NE+L+NFQS++++VARL ++C DP+
Sbjct: 237 REWIVSSVGVKMLVSDDEYFLMELTRNEILNNFQSLSQSVARLSKKCKDPM 389
>TC89130 similar to GP|12003396|gb|AAG43555.1 Avr9/Cf-9 rapidly elicited
protein 137 {Nicotiana tabacum}, partial (5%)
Length = 1159
Score = 107 bits (267), Expect = 1e-23
Identities = 64/239 (26%), Positives = 124/239 (51%), Gaps = 9/239 (3%)
Frame = +1
Query: 18 VSRKGASDNKAVVVGVLGSEVAGLMLKVVNLWQSLSDEEVLSLREGIVNSAGVRKLVSED 77
+SR+ + + +G+L + M ++++L+ SLSDEE+ L + +V S GV L S
Sbjct: 121 MSRRPKTMPEPETLGILAFDAGKTMCRLISLYSSLSDEEITKLLDEVVKSKGVTYLNSNQ 300
Query: 78 DDYLMELVLNEVLDNFQSVARTVARLGRRCVDPVYHRFEHFVRSPAQNYFQWSGWDYRGK 137
+++L+ L E L+ ++A TV+R+G +C D RF+ Q Y +
Sbjct: 301 ENFLLTLAAAERLEELDNIAVTVSRIGEKCCDLGLARFDLVYADLKQGVIDLRKLPYNSR 480
Query: 138 KMERKVKRMEKFVAAMTQLCQELEVLAEVEQTLRRMQ---------ANPVLHRGKLPEFQ 188
+ +++ EK ++A + L +E +AE+E ++ Q P L + + F
Sbjct: 481 SSIKIIEKAEKLISATSSLYSAMEYMAELEAAEKKRQQQQRYWNTTTKPSL-KPNMEYFN 657
Query: 189 KKVTLQRQEVRNLRDMSPWNRSYDYVVRLLVRSLFTILERIILVFGNNHLATKQNENES 247
+K+ QR++V+N ++ S W +++D V ++ R + + RI VFG ++ +Q+EN +
Sbjct: 658 EKLVFQRKQVQNFKETSLWKQTFDKTVGIMARLVCIVYARICSVFG-AYINEEQDENNN 831
>BG645895 weakly similar to GP|7939524|dbj contains similarity to unknown
protein~gb|AAD25764.1~gene_id:K14B15.5 {Arabidopsis
thaliana}, partial (28%)
Length = 681
Score = 95.9 bits (237), Expect = 4e-20
Identities = 50/132 (37%), Positives = 75/132 (55%)
Frame = +1
Query: 21 KGASDNKAVVVGVLGSEVAGLMLKVVNLWQSLSDEEVLSLREGIVNSAGVRKLVSEDDDY 80
K + K +G+L EVA +M K + L++SLS+ E+ LR I+NS GVR LVS ++ Y
Sbjct: 286 KRKENKKVETIGILSFEVANVMSKTIQLYKSLSESEITKLRNEILNSEGVRNLVSSEEGY 465
Query: 81 LMELVLNEVLDNFQSVARTVARLGRRCVDPVYHRFEHFVRSPAQNYFQWSGWDYRGKKME 140
L ELV E L+ VA V+RLG++C P FEH + + + K ME
Sbjct: 466 LFELVRREKLEELNRVAGVVSRLGKKCSVPALQGFEHVYGDIVSGVIDVNEFGFLVKHME 645
Query: 141 RKVKRMEKFVAA 152
V++M+++V+A
Sbjct: 646 GMVRKMDRYVSA 681
>AW329706 weakly similar to GP|7939524|dbj| contains similarity to unknown
protein~gb|AAD25764.1~gene_id:K14B15.5 {Arabidopsis
thaliana}, partial (9%)
Length = 445
Score = 53.9 bits (128), Expect = 2e-07
Identities = 38/128 (29%), Positives = 73/128 (56%), Gaps = 2/128 (1%)
Frame = +2
Query: 168 QTLRRMQANPVLHRGKLPEFQKKVTLQRQEVRNLRDMSPWNRSYDYVVRLLVRSLFTILE 227
+T+++ Q N + F++++ Q+Q+VR L+++S WN+++D VV LL R++ T+
Sbjct: 11 ETVKKFQNNSQNEESRRG-FEQRLVWQKQDVRQLKEISLWNQTFDKVVELLARTVCTLYA 187
Query: 228 RIILVFGNNHLATKQNENESPNTTANNLLRSQ--SFSVFMHSSVHPSENDLYGFHSGHLG 285
RI +VFG++ +++ N + L++++ S S ++ V+ SE L HS G
Sbjct: 188 RICMVFGDS--TVRKDGLGIGNCEGSPLVQNECGSASSLINVEVNLSEK-LKRSHSKKTG 358
Query: 286 RRLASNSG 293
L+S G
Sbjct: 359 YNLSSIGG 382
>BE942366 similar to GP|7939524|dbj| contains similarity to unknown
protein~gb|AAD25764.1~gene_id:K14B15.5 {Arabidopsis
thaliana}, partial (12%)
Length = 391
Score = 52.4 bits (124), Expect = 5e-07
Identities = 22/36 (61%), Positives = 30/36 (83%)
Frame = +3
Query: 505 KANILLVQTLYFANQVKTEAAIVDLLVGLNYVCRID 540
+ N+LL QTLYFA+++KTE AI +LL GLNY+CR +
Sbjct: 18 RTNVLLFQTLYFADKIKTEEAICELLKGLNYICRYE 125
>TC93690 similar to PIR|D86467|D86467 protein F23M19.3 [imported] -
Arabidopsis thaliana, partial (23%)
Length = 485
Score = 51.6 bits (122), Expect = 8e-07
Identities = 32/93 (34%), Positives = 46/93 (49%)
Frame = +3
Query: 400 LGDAALTLHYANMIVLIEQMVSSPHLTDPEPRDDLYKMLPATIRAVLRHKLKGRKKSKSS 459
LG A L LHYAN+++ I+ +V+ RD LY+ LP I+ LR KL S
Sbjct: 156 LGPAGLALHYANIVLQIDTLVARSISMPANTRDTLYQNLPPNIKLALRSKLP------SF 317
Query: 460 WVYNAEIAAEWSAVLAQILDWLAPLAHDTIRWH 492
V A+ + + L WL P+A +T + H
Sbjct: 318 HVVEELTVADIKDEMEKTLHWLGPIATNTSKAH 416
>TC83228 weakly similar to GP|8953735|dbj|BAA98099.1 contains similarity to
unknown protein~emb|CAB85560.1~gene_id:K10D11.2
{Arabidopsis thaliana}, partial (8%)
Length = 974
Score = 50.4 bits (119), Expect = 2e-06
Identities = 23/51 (45%), Positives = 36/51 (70%)
Frame = +3
Query: 31 VGVLGSEVAGLMLKVVNLWQSLSDEEVLSLREGIVNSAGVRKLVSEDDDYL 81
V VL E+AG+M K+++L+QSLSD ++ +R V GVRK++S D+ +L
Sbjct: 384 VAVLAFEIAGVMSKLLHLFQSLSDATIVRIRNDAVTLEGVRKIISNDESFL 536
>TC93869 weakly similar to GP|20466802|gb|AAM20718.1 unknown protein
{Arabidopsis thaliana}, partial (29%)
Length = 686
Score = 50.1 bits (118), Expect = 2e-06
Identities = 45/183 (24%), Positives = 72/183 (38%), Gaps = 10/183 (5%)
Frame = +1
Query: 73 LVSEDDDYLMELVLNEVLDNFQSVARTVARLGRRCVDPVYHRFEHFVRSPAQNYFQWSGW 132
LVS D L+ V + + F + +R VAR G C DP +H + YF +
Sbjct: 1 LVSTDTKELISFVEADKREEFNAFSREVARFGNICKDPQWHNLD--------RYFSRLDF 156
Query: 133 DYRGKKM-----ERKVKRMEKFVAAMTQLCQELEVLAEVEQTLRRMQ-----ANPVLHRG 182
D K E+ V+ + +L EL L +Q + N L+
Sbjct: 157 DALSNKQPRVEAEKTVQDLSSLAQNTAELYHELNALDRFQQDYNQKVKELEFLNLPLNGE 336
Query: 183 KLPEFQKKVTLQRQEVRNLRDMSPWNRSYDYVVRLLVRSLFTILERIILVFGNNHLATKQ 242
L F ++ QR+ V++L+ S W++ + +V LV + I G N +
Sbjct: 337 GLAAFHSELKHQRKLVKSLQRKSLWSKHLEEIVEKLVEVATHTHQAIFEFLGKNGKIAVK 516
Query: 243 NEN 245
N N
Sbjct: 517 NRN 525
Score = 46.6 bits (109), Expect = 3e-05
Identities = 27/61 (44%), Positives = 34/61 (55%)
Frame = +1
Query: 386 KLSMSNRLKPASFTLGDAALTLHYANMIVLIEQMVSSPHLTDPEPRDDLYKMLPATIRAV 445
K+++ NR P LG+A L LHYAN+I I + S P + P RD LYK LP I
Sbjct: 502 KIAVKNRNGPER--LGEAGLALHYANIINQINVIASRPTILPPNMRDTLYKGLPNNIXNA 675
Query: 446 L 446
L
Sbjct: 676 L 678
>TC91715 weakly similar to GP|20466802|gb|AAM20718.1 unknown protein
{Arabidopsis thaliana}, partial (26%)
Length = 744
Score = 47.0 bits (110), Expect = 2e-05
Identities = 24/52 (46%), Positives = 32/52 (61%)
Frame = +3
Query: 400 LGDAALTLHYANMIVLIEQMVSSPHLTDPEPRDDLYKMLPATIRAVLRHKLK 451
LG+A L LHYANMI I + S P P RD LY+ LP +I++ L +L+
Sbjct: 588 LGEAGLALHYANMINQIYMIASRPASLPPNTRDTLYQGLPNSIKSALPSRLQ 743
Score = 32.0 bits (71), Expect = 0.66
Identities = 26/109 (23%), Positives = 50/109 (45%), Gaps = 5/109 (4%)
Frame = +3
Query: 140 ERKVKRMEKFVAAMTQLCQELEVLA----EVEQTLRRMQA-NPVLHRGKLPEFQKKVTLQ 194
E+ ++ V +L EL + +Q ++ M++ N L + FQ ++ Q
Sbjct: 243 EKTMQEFTSLVHHTAELYHELSAYERFQHDYQQKIKEMESLNLPLKGESITIFQSELKHQ 422
Query: 195 RQEVRNLRDMSPWNRSYDYVVRLLVRSLFTILERIILVFGNNHLATKQN 243
++ V NL+ S W+R + +V LV + I + I + GN+ +N
Sbjct: 423 KKLVTNLKKKSLWSRYLEEIVEKLVDIVTYIHQAIRELLGNHGTGAVKN 569
>BQ122396 similar to GP|7939524|dbj| contains similarity to unknown
protein~gb|AAD25764.1~gene_id:K14B15.5 {Arabidopsis
thaliana}, partial (7%)
Length = 498
Score = 38.5 bits (88), Expect = 0.007
Identities = 16/29 (55%), Positives = 24/29 (82%)
Frame = +2
Query: 397 SFTLGDAALTLHYANMIVLIEQMVSSPHL 425
S TLG +AL LHYAN+I++I++++ PHL
Sbjct: 407 SSTLGGSALALHYANVIIVIKKLLRYPHL 493
>BQ137291 similar to GP|13605829|gb| AT5g04550/T32M21_140 {Arabidopsis
thaliana}, partial (3%)
Length = 985
Score = 33.1 bits (74), Expect = 0.30
Identities = 15/29 (51%), Positives = 23/29 (78%)
Frame = +1
Query: 31 VGVLGSEVAGLMLKVVNLWQSLSDEEVLS 59
V VL E+AG+M K+++L+QSLSD ++S
Sbjct: 85 VAVLAFEIAGVMSKLLHLFQSLSDATIVS 171
>TC84009 similar to GP|13548324|emb|CAC35871. putative protein {Arabidopsis
thaliana}, partial (12%)
Length = 737
Score = 32.7 bits (73), Expect = 0.39
Identities = 18/50 (36%), Positives = 30/50 (60%)
Frame = +3
Query: 36 SEVAGLMLKVVNLWQSLSDEEVLSLREGIVNSAGVRKLVSEDDDYLMELV 85
S + L+ K +L +SLS + + L+E ++ S V LVS+D D L+ +V
Sbjct: 549 SRLQTLLSKXFHLLESLSTKSIRHLKEEVLLSETVHDLVSKDKDELLTIV 698
>TC91357 similar to GP|2194137|gb|AAB61112.1| ESTs gb|R29947 gb|H76702 come
from this gene. {Arabidopsis thaliana}, partial (40%)
Length = 766
Score = 32.0 bits (71), Expect = 0.66
Identities = 22/80 (27%), Positives = 35/80 (43%), Gaps = 6/80 (7%)
Frame = +3
Query: 227 ERIILVFG------NNHLATKQNENESPNTTANNLLRSQSFSVFMHSSVHPSENDLYGFH 280
E+ I++FG NNHL T + NTT NN R + + P D H
Sbjct: 285 EKSIMIFGVRLTGGNNHLNTT-----TTNTTINNSFRKSASMTNLSQYEQPPPQDSNPAH 449
Query: 281 SGHLGRRLASNSGILVDKKQ 300
+G++ + SG ++K+
Sbjct: 450 AGYVSHDIVHASGRSRERKR 509
>TC90357 similar to GP|20466802|gb|AAM20718.1 unknown protein {Arabidopsis
thaliana}, partial (6%)
Length = 736
Score = 32.0 bits (71), Expect = 0.66
Identities = 13/46 (28%), Positives = 31/46 (67%)
Frame = +2
Query: 491 WHSVRSFEKEHATLKANILLVQTLYFANQVKTEAAIVDLLVGLNYV 536
W + + ++ T ++N + +QTLY+A++ K + I++LLV ++++
Sbjct: 50 WANTSNDFGDNTTKESNPIRLQTLYYADKQKIDVYIIELLVWIHHL 187
>TC86258 similar to GP|16930433|gb|AAL31902.1 AT4g17230/dl4650c {Arabidopsis
thaliana}, partial (62%)
Length = 2237
Score = 32.0 bits (71), Expect = 0.66
Identities = 26/82 (31%), Positives = 35/82 (41%), Gaps = 15/82 (18%)
Frame = -1
Query: 380 RIRIYSKLSM----SNRLKP--------ASFTLGDAALTLHYANMIVLIEQMVSSP---H 424
RI IY KL +N LK + F G T+HYAN VL++Q+ + H
Sbjct: 2162 RIDIYKKLRFCRLHNNLLKGTCWK*S*ISKFIYGKTLDTIHYANTKVLVKQVTKNKGQLH 1983
Query: 425 LTDPEPRDDLYKMLPATIRAVL 446
LT P R P I + +
Sbjct: 1982 LTHPSRRSGHSSFHPTKIESTI 1917
>BE325721 similar to SP|P49147|PA2Y_ Cytosolic phospholipase A2 (CPLA2)
[Includes: Phospholipase A2 (EC 3.1.1.4) (CPLA2),
partial (2%)
Length = 654
Score = 30.0 bits (66), Expect = 2.5
Identities = 12/31 (38%), Positives = 19/31 (60%)
Frame = +1
Query: 220 RSLFTILERIILVFGNNHLATKQNENESPNT 250
+S F L +I++FG+ H K N+N +P T
Sbjct: 22 KSTFFSLSLLIIIFGSKHGKRKPNQNSNPRT 114
>TC86402 homologue to GP|15148926|gb|AAK84890.1 TGA-type basic leucine zipper
protein TGA2.2 {Phaseolus vulgaris}, partial (96%)
Length = 1970
Score = 29.3 bits (64), Expect = 4.3
Identities = 24/101 (23%), Positives = 46/101 (44%), Gaps = 1/101 (0%)
Frame = +3
Query: 83 ELVLNEVLDNFQSVARTVARL-GRRCVDPVYHRFEHFVRSPAQNYFQWSGWDYRGKKMER 141
++ L ++DNF + + RL G V+H ++PA+ F W G G +
Sbjct: 912 DIELRTIVDNFMTQFEDIYRLKGVAAKADVFHILSGMWKTPAERCFMWIG----GFRSSE 1079
Query: 142 KVKRMEKFVAAMTQLCQELEVLAEVEQTLRRMQANPVLHRG 182
+K + + +T+ Q+L + ++Q+ QA L +G
Sbjct: 1080 LLKLLVSHLEPLTE--QQLMGIYNLQQS--SQQAEDALSQG 1190
>CB892620 weakly similar to GP|22655012|g At1g73080/F3N23_28 {Arabidopsis
thaliana}, partial (9%)
Length = 704
Score = 28.5 bits (62), Expect = 7.3
Identities = 21/83 (25%), Positives = 36/83 (43%), Gaps = 13/83 (15%)
Frame = +1
Query: 169 TLRRMQANPVLHRGKLPEFQKKVTLQRQEVR-------------NLRDMSPWNRSYDYVV 215
TLRR+ N G LP+F + L+ ++ N +++ N S +
Sbjct: 133 TLRRLFLNQNNFTGSLPDFASNLNLKYMDISKNNISGPIPSSLGNCTNLTYINLSRNKFA 312
Query: 216 RLLVRSLFTILERIILVFGNNHL 238
RL+ L +L +IL +N+L
Sbjct: 313 RLIPSELGNLLNLVILELSHNNL 381
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,461,721
Number of Sequences: 36976
Number of extensions: 209909
Number of successful extensions: 1075
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 1063
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1070
length of query: 572
length of database: 9,014,727
effective HSP length: 101
effective length of query: 471
effective length of database: 5,280,151
effective search space: 2486951121
effective search space used: 2486951121
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0378b.5


