

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
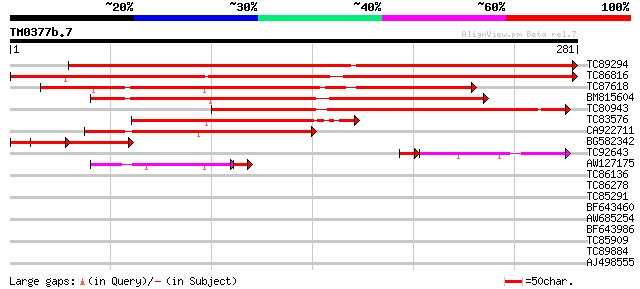
Query= TM0377b.7
(281 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89294 similar to GP|10176982|dbj|BAB10214. contains similarity... 444 e-125
TC86816 similar to GP|10176982|dbj|BAB10214. contains similarity... 385 e-107
TC87618 similar to GP|10176982|dbj|BAB10214. contains similarity... 215 2e-56
BM815604 similar to GP|10176982|db contains similarity to unknow... 213 7e-56
TC80943 similar to GP|15451150|gb|AAK96846.1 Unknown protein {Ar... 194 3e-50
TC83576 similar to GP|15293013|gb|AAK93617.1 unknown protein {Ar... 108 2e-24
CA922711 similar to GP|20197959|gb| expressed protein {Arabidops... 103 7e-23
BG582342 65 3e-11
TC92643 similar to GP|15293013|gb|AAK93617.1 unknown protein {Ar... 47 7e-07
AW127175 similar to GP|15293013|gb| unknown protein {Arabidopsis... 45 7e-06
TC86136 weakly similar to PIR|A96673|A96673 probable cytochrome ... 30 1.0
TC86278 similar to GP|17473703|gb|AAL38307.1 germin-like protein... 29 1.8
TC85291 similar to SP|Q43070|GAE1_PEA UDP-glucose 4-epimerase (E... 29 2.3
BF643460 similar to PIR|T09640|T09 protein phosphatase 2C - alfa... 28 3.0
AW685254 similar to GP|7270274|emb WD-repeat protein-like protei... 28 3.0
BF643986 weakly similar to PIR|T02483|T024 probable protein phos... 28 3.0
TC85909 similar to GP|13277342|emb|CAC34417. Germin-like protein... 28 3.9
TC89884 similar to GP|2459431|gb|AAB80666.1| unknown protein {Ar... 28 3.9
AJ498555 weakly similar to SP|Q9FMA8|GL1B Germin-like protein su... 27 6.7
>TC89294 similar to GP|10176982|dbj|BAB10214. contains similarity to unknown
protein~emb|CAB89322.1~gene_id:MYH19.8 {Arabidopsis
thaliana}, partial (82%)
Length = 838
Score = 444 bits (1143), Expect = e-125
Identities = 205/252 (81%), Positives = 232/252 (91%)
Frame = +3
Query: 30 RNRRRQKKMPPVQKLFDTCKEVFASGGTGFIPPPQDIQRLQAVLDAIRPEDVGLKPDMPH 89
RNRRRQKKMPPVQKLF+TCKEVFAS GTG +PP +DI +L++VLD I+PEDV L P+MP+
Sbjct: 3 RNRRRQKKMPPVQKLFETCKEVFASSGTGIVPPAEDIDKLRSVLDGIKPEDVDLDPNMPY 182
Query: 90 FRSSSAQRIPKITYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSY 149
FR++++ R PKITYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSY
Sbjct: 183 FRANASHRRPKITYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSY 362
Query: 150 DWVVDLPPESPTIVKPTENQALEMRLAKVKVDADFTAPCNPSILYPEDGGNMHCFTAVTA 209
DWVVDLPPESPTI KP+E+ ++RLAKVKVD DFTAPCNPSILYPEDGGN+HCFTAVTA
Sbjct: 363 DWVVDLPPESPTIFKPSESP--DLRLAKVKVDDDFTAPCNPSILYPEDGGNLHCFTAVTA 536
Query: 210 CAVLDVLGPPYSDYEGRHCTYYNNYPFSDISVEGISIPEEERNGYEWLQEKEQLEDLEVD 269
CAVLDVLGPPYSD++GRHCTYY NYPFS+ VEG+SIPEEER+ YEWLQEK+QLEDL+V+
Sbjct: 537 CAVLDVLGPPYSDFDGRHCTYYTNYPFSNFQVEGLSIPEEERSVYEWLQEKDQLEDLKVE 716
Query: 270 GKMYSGPKIHES 281
G+MYSGP I ES
Sbjct: 717 GRMYSGPTIVES 752
>TC86816 similar to GP|10176982|dbj|BAB10214. contains similarity to unknown
protein~emb|CAB89322.1~gene_id:MYH19.8 {Arabidopsis
thaliana}, partial (71%)
Length = 1314
Score = 385 bits (988), Expect = e-107
Identities = 189/285 (66%), Positives = 222/285 (77%), Gaps = 4/285 (1%)
Frame = +3
Query: 1 MGIERTLADRKGRSFCELPRETITSS----GSKRNRRRQKKMPPVQKLFDTCKEVFASGG 56
MGIER + +RKGR CEL ET T++ ++R+ RR+ +M PVQKLF CK VFA+
Sbjct: 276 MGIERNMVERKGRELCELLDETNTNNMKPRKNRRHLRRRTEMTPVQKLFLACKHVFANAA 455
Query: 57 TGFIPPPQDIQRLQAVLDAIRPEDVGLKPDMPHFRSSSAQRIPKITYLHIYECEKFSMGI 116
G +P Q I+ L++VL I+PED+GLKPDMP+F + + PKITYLHIYECEKFSMGI
Sbjct: 456 HGIVPSSQHIEMLRSVLAGIKPEDLGLKPDMPYFSNINGGT-PKITYLHIYECEKFSMGI 632
Query: 117 FCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSYDWVVDLPPESPTIVKPTENQALEMRLA 176
FCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSYDW DLP + ++ Q E RLA
Sbjct: 633 FCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSYDWAGDLPADV------SQTQIPEKRLA 794
Query: 177 KVKVDADFTAPCNPSILYPEDGGNMHCFTAVTACAVLDVLGPPYSDYEGRHCTYYNNYPF 236
K+KVDADFTAPCNPSILYP+DGGNMHCFTAVTACAVLDVLGPPYSD +GRHC YY ++PF
Sbjct: 795 KIKVDADFTAPCNPSILYPDDGGNMHCFTAVTACAVLDVLGPPYSDPDGRHCAYYRSFPF 974
Query: 237 SDISVEGISIPEEERNGYEWLQEKEQLEDLEVDGKMYSGPKIHES 281
S+ VEGISIPEEE+ YEWLQE+E+ E L+V KMYS K E+
Sbjct: 975 SNFPVEGISIPEEEKKDYEWLQEREKPESLQVIVKMYSSSKTMEN 1109
>TC87618 similar to GP|10176982|dbj|BAB10214. contains similarity to unknown
protein~emb|CAB89322.1~gene_id:MYH19.8 {Arabidopsis
thaliana}, partial (46%)
Length = 1037
Score = 215 bits (547), Expect = 2e-56
Identities = 109/219 (49%), Positives = 146/219 (65%), Gaps = 3/219 (1%)
Frame = +2
Query: 16 CELPRETITSSGSKRNRRRQKKMPP--VQKLFDTCKEVFASGGTGFIPPPQDIQRLQAVL 73
C + R N RR KK P +Q+LFD+CK+ F GT +P P+D+ +L +L
Sbjct: 404 CYVKRVIAKKRNKPYNHRRVKKHVPKALQELFDSCKQTFKGPGT--VPSPRDVHKLCHIL 577
Query: 74 DAIRPEDVGLKPDMPHFRSSSA-QRIPKITYLHIYECEKFSMGIFCLPPSGVIPLHNHPG 132
D ++PEDVGL D+ F+ + + ++TY +Y+C+ FS+ IF LP GVIPLHNHPG
Sbjct: 578 DNMKPEDVGLSRDLQFFKPGNIIKENQRVTYTTVYKCDNFSLCIFFLPERGVIPLHNHPG 757
Query: 133 MTVFSKLLFGTMHIKSYDWVVDLPPESPTIVKPTENQALEMRLAKVKVDADFTAPCNPSI 192
MTVFSKLL G MHIKSYDWV S +++P+ ++RLAK+K + FTAPC+ S+
Sbjct: 758 MTVFSKLLLGQMHIKSYDWVDH--EASHNLLQPSS----KLRLAKLKANKTFTAPCDTSV 919
Query: 193 LYPEDGGNMHCFTAVTACAVLDVLGPPYSDYEGRHCTYY 231
LYP GGN+H FTA+T CAVLDV+GPPYS +GR C+YY
Sbjct: 920 LYPTTGGNIHEFTAITPCAVLDVIGPPYSKEDGRDCSYY 1036
>BM815604 similar to GP|10176982|db contains similarity to unknown
protein~emb|CAB89322.1~gene_id:MYH19.8 {Arabidopsis
thaliana}, partial (45%)
Length = 793
Score = 213 bits (542), Expect = 7e-56
Identities = 104/198 (52%), Positives = 138/198 (69%), Gaps = 1/198 (0%)
Frame = +2
Query: 41 VQKLFDTCKEVFASGGTGFIPPPQDIQRLQAVLDAIRPEDVGLKPDMPHFRSSSAQRI-P 99
+QKLF +CKE F T +P PQ++ +L +LD ++PEDVGL D+ F+ S R P
Sbjct: 110 LQKLFVSCKETFKGPNT--VPSPQNVHKLCHILDNMKPEDVGLSKDLQFFKPESIFRENP 283
Query: 100 KITYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSYDWVVDLPPES 159
++TY IY+C+ FS+ IF LP GVIPLHNHPGMTVFSKLL G MHIKSYDWV
Sbjct: 284 RVTYTTIYKCDNFSLCIFFLPSKGVIPLHNHPGMTVFSKLLLGQMHIKSYDWV------D 445
Query: 160 PTIVKPTENQALEMRLAKVKVDADFTAPCNPSILYPEDGGNMHCFTAVTACAVLDVLGPP 219
P + Q ++RLAK+K + FT+PC+ S+LYP+ GGN+H FTA+T CAVLDV+GPP
Sbjct: 446 PDVSHNLLKQPSQLRLAKLKANKVFTSPCDTSVLYPKTGGNIHEFTAITPCAVLDVIGPP 625
Query: 220 YSDYEGRHCTYYNNYPFS 237
YS +GR C+YY ++ ++
Sbjct: 626 YSKDDGRDCSYYRDHLYT 679
>TC80943 similar to GP|15451150|gb|AAK96846.1 Unknown protein {Arabidopsis
thaliana}, partial (55%)
Length = 661
Score = 194 bits (493), Expect = 3e-50
Identities = 93/178 (52%), Positives = 123/178 (68%)
Frame = +2
Query: 101 ITYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSYDWVVDLPPESP 160
ITY+ I+E + F+M +FC P S VIPLH+HP MTVFSKLL+G++H+K+YDWV E P
Sbjct: 44 ITYVDIHESDSFTMCMFCFPTSSVIPLHDHPQMTVFSKLLYGSLHVKAYDWV-----EPP 208
Query: 161 TIVKPTENQALEMRLAKVKVDADFTAPCNPSILYPEDGGNMHCFTAVTACAVLDVLGPPY 220
IVK ++RLAK+ VD APC S+LYP GGN+HCFTAVT CA+LDVL PPY
Sbjct: 209 CIVKSKGPGHAQVRLAKLAVDKVLNAPCETSVLYPNCGGNIHCFTAVTPCAMLDVLAPPY 388
Query: 221 SDYEGRHCTYYNNYPFSDISVEGISIPEEERNGYEWLQEKEQLEDLEVDGKMYSGPKI 278
+YEGR CTYY++YP+S S S+ + + + Y WL E E +L ++ +Y+GP I
Sbjct: 389 KEYEGRKCTYYHDYPYSTFSAGNGSLCDGDEDEYAWLAEVEP-SNLYMNSGVYAGPAI 559
>TC83576 similar to GP|15293013|gb|AAK93617.1 unknown protein {Arabidopsis
thaliana}, partial (37%)
Length = 695
Score = 108 bits (270), Expect = 2e-24
Identities = 56/121 (46%), Positives = 80/121 (65%), Gaps = 8/121 (6%)
Frame = +3
Query: 61 PPPQDIQRLQAVLDAIRPEDVGLKPDMPHFRSSSAQ--------RIPKITYLHIYECEKF 112
P + I ++ L+ ++P DVGL+ + R+ + Q + P I YLH++EC+ F
Sbjct: 333 PCRKAISKVCEKLEKMKPSDVGLEQEAQVVRTWNGQMPESNGNHQPPPIKYLHLHECDSF 512
Query: 113 SMGIFCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSYDWVVDLPPESPTIVKPTENQALE 172
S+GIFC+P S VIPLHNHPGMTV SKL++GT+H+KSYDW +D P PT PTE +A +
Sbjct: 513 SIGIFCMPTSSVIPLHNHPGMTVLSKLIYGTVHVKSYDW-IDFP--GPT--DPTEARAAK 677
Query: 173 M 173
+
Sbjct: 678 L 680
>CA922711 similar to GP|20197959|gb| expressed protein {Arabidopsis
thaliana}, partial (21%)
Length = 806
Score = 103 bits (257), Expect = 7e-23
Identities = 47/123 (38%), Positives = 77/123 (62%), Gaps = 8/123 (6%)
Frame = +1
Query: 38 MPPVQKLFDTCKEVFASGGTGFIPPPQDIQRLQAVLDAIRPEDVGLKPDMPHFRS----- 92
MP +QKL+D CK + G + +++++ VLD ++P VGL + RS
Sbjct: 442 MPYIQKLYDVCKASLSPEGP---ISEEALEKVRTVLDGLKPSHVGLDHEAQLARSWKRSM 612
Query: 93 ---SSAQRIPKITYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSY 149
+ P I Y+H++EC+KFS+G+FC+ P +IPLH+HP MTV SK L+G++H++++
Sbjct: 613 NGGNGTPEGPPIKYIHLHECDKFSIGVFCMSPGSLIPLHDHPRMTVLSKGLYGSLHVEAF 792
Query: 150 DWV 152
DW+
Sbjct: 793 DWI 801
>BG582342
Length = 651
Score = 65.1 bits (157), Expect = 3e-11
Identities = 31/51 (60%), Positives = 36/51 (69%)
Frame = +2
Query: 11 KGRSFCELPRETITSSGSKRNRRRQKKMPPVQKLFDTCKEVFASGGTGFIP 61
K F R+ + ++G NRRRQKKMPPVQKLF+TCKEVFAS GTG P
Sbjct: 416 KEEIFVNCQRKQLLAAGQGGNRRRQKKMPPVQKLFETCKEVFASSGTGNCP 568
Score = 56.2 bits (134), Expect = 1e-08
Identities = 26/30 (86%), Positives = 28/30 (92%)
Frame = +1
Query: 1 MGIERTLADRKGRSFCELPRETITSSGSKR 30
MGIERTLADRKGR FCELP+ETITSS S+R
Sbjct: 385 MGIERTLADRKGRDFCELPKETITSSRSRR 474
>TC92643 similar to GP|15293013|gb|AAK93617.1 unknown protein {Arabidopsis
thaliana}, partial (7%)
Length = 648
Score = 46.6 bits (109), Expect(2) = 7e-07
Identities = 28/82 (34%), Positives = 41/82 (49%), Gaps = 7/82 (8%)
Frame = +2
Query: 204 FTAVTACAVLDVLGPPYS---DYEGRHCTYYNNYPFSDISV----EGISIPEEERNGYEW 256
F A+T CA+ D+L PPYS + GR+C+Y+ +D+ V G+S E W
Sbjct: 32 FKAITPCALFDILTPPYSLEEEVNGRNCSYFRKSLRTDLPVLEELRGMSSSE-----ITW 196
Query: 257 LQEKEQLEDLEVDGKMYSGPKI 278
L++ DL + Y GP I
Sbjct: 197 LEKIPPPSDLIIGNGQYRGPNI 262
Score = 23.5 bits (49), Expect(2) = 7e-07
Identities = 7/10 (70%), Positives = 8/10 (80%)
Frame = +3
Query: 194 YPEDGGNMHC 203
YP GGN+HC
Sbjct: 3 YPNRGGNLHC 32
>AW127175 similar to GP|15293013|gb| unknown protein {Arabidopsis thaliana},
partial (37%)
Length = 452
Score = 44.7 bits (104), Expect(2) = 7e-06
Identities = 26/82 (31%), Positives = 44/82 (52%), Gaps = 10/82 (12%)
Frame = +3
Query: 41 VQKLFDTCKEVFASGGTGFIPPPQDI-QRLQAVLDAIRPEDVGLKPDMPHFRSSSA---- 95
VQ+L+ CK F+ G P +++ ++++ LD I+P DVGL+ + R+ S
Sbjct: 192 VQRLYRLCKASFSPDG----PVSEEVVKKVREKLDRIKPSDVGLEQEAQVVRNMSRTVLE 359
Query: 96 -----QRIPKITYLHIYECEKF 112
+P I YLH++EC+ F
Sbjct: 360 QNGSHHSLPAIKYLHLHECDSF 425
Score = 21.9 bits (45), Expect(2) = 7e-06
Identities = 7/9 (77%), Positives = 9/9 (99%)
Frame = +1
Query: 112 FSMGIFCLP 120
FS+GIFC+P
Sbjct: 424 FSIGIFCMP 450
>TC86136 weakly similar to PIR|A96673|A96673 probable cytochrome P450
F13O11.25 [imported] - Arabidopsis thaliana, partial
(26%)
Length = 721
Score = 30.0 bits (66), Expect = 1.0
Identities = 22/64 (34%), Positives = 31/64 (48%), Gaps = 2/64 (3%)
Frame = +2
Query: 105 HIYECEKFSMGIFCLPPSGVIPLHNH-PGMTVFSKLLFGTMHIKSY-DWVVDLPPESPTI 162
HIY+ + S G NH P + +FS+L M + S V+DLP SPT+
Sbjct: 158 HIYQSSQASNGF-----------ENHSPSLNLFSELFMPNMVLSSILISVLDLPFSSPTV 304
Query: 163 VKPT 166
+ PT
Sbjct: 305 LWPT 316
>TC86278 similar to GP|17473703|gb|AAL38307.1 germin-like protein
{Arabidopsis thaliana}, partial (88%)
Length = 908
Score = 29.3 bits (64), Expect = 1.8
Identities = 11/14 (78%), Positives = 12/14 (85%)
Frame = +2
Query: 119 LPPSGVIPLHNHPG 132
L P+GVIPLH HPG
Sbjct: 314 LGPAGVIPLHTHPG 355
>TC85291 similar to SP|Q43070|GAE1_PEA UDP-glucose 4-epimerase (EC 5.1.3.2)
(Galactowaldenase) (UDP- galactose 4-epimerase). [Garden
pea], partial (78%)
Length = 1018
Score = 28.9 bits (63), Expect = 2.3
Identities = 15/58 (25%), Positives = 28/58 (47%)
Frame = +1
Query: 13 RSFCELPRETITSSGSKRNRRRQKKMPPVQKLFDTCKEVFASGGTGFIPPPQDIQRLQ 70
+S L ++ S N + P+QK+ + +++ +GG+GFI +Q LQ
Sbjct: 94 KSIRNLKPTKVSP*SSSLNLSLSLSLVPIQKMVSSSQKILVTGGSGFIGTHTVLQLLQ 267
>BF643460 similar to PIR|T09640|T09 protein phosphatase 2C - alfalfa, partial
(33%)
Length = 565
Score = 28.5 bits (62), Expect = 3.0
Identities = 12/26 (46%), Positives = 18/26 (69%), Gaps = 1/26 (3%)
Frame = -3
Query: 87 MPHFRSSSAQRIPKITYLHIYE-CEK 111
+ H SS RIPK+ YLH+++ C+K
Sbjct: 398 LTHSFPSSVPRIPKLLYLHVHQICQK 321
>AW685254 similar to GP|7270274|emb WD-repeat protein-like protein
{Arabidopsis thaliana}, partial (35%)
Length = 657
Score = 28.5 bits (62), Expect = 3.0
Identities = 15/37 (40%), Positives = 21/37 (56%)
Frame = -1
Query: 104 LHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLL 140
L + EC+K S PP ++PL NHPG + +LL
Sbjct: 426 LLLLECQKHSKTSRFHPPQ-LLPLCNHPGSAPYEELL 319
>BF643986 weakly similar to PIR|T02483|T024 probable protein phosphatase 2C
At2g30020 [imported] - Arabidopsis thaliana, partial
(16%)
Length = 448
Score = 28.5 bits (62), Expect = 3.0
Identities = 12/26 (46%), Positives = 18/26 (69%), Gaps = 1/26 (3%)
Frame = -1
Query: 87 MPHFRSSSAQRIPKITYLHIYE-CEK 111
+ H SS RIPK+ YLH+++ C+K
Sbjct: 418 LTHSFPSSVPRIPKLLYLHVHQICQK 341
>TC85909 similar to GP|13277342|emb|CAC34417. Germin-like protein {Pisum
sativum}, partial (95%)
Length = 1551
Score = 28.1 bits (61), Expect = 3.9
Identities = 12/26 (46%), Positives = 16/26 (61%)
Frame = +2
Query: 119 LPPSGVIPLHNHPGMTVFSKLLFGTM 144
L GVIPLH HPG + ++ GT+
Sbjct: 296 LAAGGVIPLHTHPGASEVLVVIQGTI 373
>TC89884 similar to GP|2459431|gb|AAB80666.1| unknown protein {Arabidopsis
thaliana}, partial (13%)
Length = 878
Score = 28.1 bits (61), Expect = 3.9
Identities = 15/50 (30%), Positives = 26/50 (52%)
Frame = +3
Query: 98 IPKITYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLLFGTMHIK 147
+P ++ I+ C + +FCL S + LH+HP + LL +H+K
Sbjct: 375 LPDLSQWKIFSC---FLDLFCLLES--LSLHHHPPKFLLVLLLLSCLHLK 509
>AJ498555 weakly similar to SP|Q9FMA8|GL1B Germin-like protein subfamily 1
member 11 precursor. [Mouse-ear cress] {Arabidopsis
thaliana}, partial (65%)
Length = 499
Score = 27.3 bits (59), Expect = 6.7
Identities = 11/29 (37%), Positives = 16/29 (54%)
Frame = +2
Query: 121 PSGVIPLHNHPGMTVFSKLLFGTMHIKSY 149
P G+ P H HP T +L GT+++ Y
Sbjct: 260 PKGINPPHTHPRATEILMVLDGTLNVGFY 346
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.138 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,225,834
Number of Sequences: 36976
Number of extensions: 164419
Number of successful extensions: 738
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 720
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 722
length of query: 281
length of database: 9,014,727
effective HSP length: 95
effective length of query: 186
effective length of database: 5,502,007
effective search space: 1023373302
effective search space used: 1023373302
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0377b.7


