

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0375a.8
(606 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
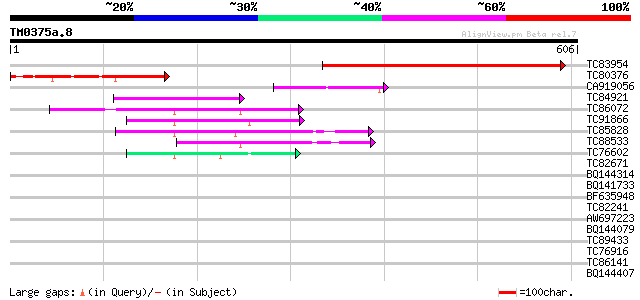
Score E
Sequences producing significant alignments: (bits) Value
TC83954 similar to SP|Q9ZSS6|THD1_ARATH Threonine dehydratase bi... 416 e-116
TC80376 similar to GP|13324801|gb|AAK18849.1 putative threonine ... 191 5e-49
CA919056 similar to PIR|T04211|T04 probable threonine dehydratas... 70 2e-12
TC84921 similar to PIR|T04211|T04211 probable threonine dehydrat... 69 7e-12
TC86072 similar to GP|11559260|dbj|BAB18760. beta-cyanoalanine s... 65 8e-11
TC91866 similar to SP|O22682|CYSN_ARATH Probable cysteine syntha... 56 3e-08
TC85828 homologue to GP|18252506|gb|AAL66291.1 cysteine synthase... 52 5e-07
TC88533 similar to GP|12081919|dbj|BAB20862. plastidic cysteine ... 47 2e-05
TC76602 similar to SP|Q43317|CYSK_CITLA Cysteine synthase (EC 4.... 45 1e-04
TC82671 similar to GP|8778327|gb|AAF79336.1| F14J16.13 {Arabidop... 37 0.022
BQ144314 weakly similar to GP|21322711|em pherophorin-dz1 protei... 33 0.41
BQ141733 homologue to GP|20161357|dbj putative RNA helicase {Ory... 32 0.92
BF635948 similar to GP|21622312|emb putative protein {Neurospora... 32 0.92
TC82241 similar to PIR|B84620|B84620 hypothetical protein At2g23... 31 1.2
AW697223 similar to GP|9757890|dbj phosphoribosylanthranilate tr... 31 1.2
BQ144079 similar to GP|6523547|emb| hydroxyproline-rich glycopro... 31 1.2
TC89433 similar to GP|13277220|emb|CAC34409. ZF-HD homeobox prot... 31 1.6
TC76916 MtN4 31 1.6
TC86141 similar to GP|20260580|gb|AAM13188.1 unknown protein {Ar... 30 2.0
BQ144407 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 30 2.0
>TC83954 similar to SP|Q9ZSS6|THD1_ARATH Threonine dehydratase biosynthetic
chloroplast precursor (EC 4.2.1.16) (Threonine
deaminase) (TD)., partial (42%)
Length = 782
Score = 416 bits (1068), Expect = e-116
Identities = 203/260 (78%), Positives = 233/260 (89%)
Frame = +2
Query: 335 GVAVKEVGEETFRLCKDLIDGVVLVNRDSICASIKDMFEENRSILEPSGALALAGAEAYC 394
GVAVKEVGEETFRLCK+L+DGVVLV+RD+ICASIKDMFEE RSILEP+GALALAGAEAYC
Sbjct: 2 GVAVKEVGEETFRLCKELVDGVVLVSRDAICASIKDMFEEKRSILEPAGALALAGAEAYC 181
Query: 395 KYYGIKGGNVVAITSGANMNFDKLRVVTELANVGRKQEAVLATVMPEEPGSFKQFCELVG 454
KYYG+KG NV+AITSGANMNFDKLR+VTELANVGRKQEA+L TVMPEEPGSFKQFCELVG
Sbjct: 182 KYYGLKGENVIAITSGANMNFDKLRIVTELANVGRKQEALLLTVMPEEPGSFKQFCELVG 361
Query: 455 QANITEFTYRFTSSDKAVVLYSVGVHTVSDLSAMKERMKSSHLKTHDLTENDLVKDHLRY 514
Q NITEF YR++S +KAVVLYSVGVHT +L M+ERM++S L TH+L+ DLVKDHLR+
Sbjct: 362 QMNITEFKYRYSSKEKAVVLYSVGVHTPLELKEMQERMETSKLVTHNLSNVDLVKDHLRH 541
Query: 515 MMGGRSDVENEVLCRFIFPERPGALMKFLDSFSPRWNISLFHYRGEGETRANILVGIQVP 574
MMG D++NE LCRF FP+RPGAL KFL+SFSPR NISLFHYR +GET AN+LVGIQVP
Sbjct: 542 MMGAPPDIQNEGLCRFTFPKRPGALTKFLNSFSPRRNISLFHYRAQGETGANVLVGIQVP 721
Query: 575 RTEMDEFHERANRLGYEYKV 594
+EM+EF++RA LGY+Y V
Sbjct: 722 SSEMEEFNDRATGLGYDYNV 781
>TC80376 similar to GP|13324801|gb|AAK18849.1 putative threonine
dehydratase/deaminase {Oryza sativa}, partial (10%)
Length = 585
Score = 191 bits (486), Expect = 5e-49
Identities = 106/176 (60%), Positives = 127/176 (71%), Gaps = 6/176 (3%)
Frame = +1
Query: 1 MAALRFSTTTHPHHPSPPPLLRRHAFHKMPTNVLHPNPRRKPFI---AATISKPAEITPS 57
M ALR S P P LLR H H+M LHPNPR KPF+ AA +SKP E+
Sbjct: 79 MEALRLS------QPQPHLLLRNHR-HRMQYTTLHPNPRFKPFVTTTAAAVSKPVEMVSV 237
Query: 58 PSSSSAAVGDHLSSPRLRVSPDSLQYEPGYLGAVPDRSRSDGGSDSDAMLTAMA---NIL 114
P S A ++SPRL+VSPDSLQY PGY+GAVPDRSR G + D ++AM +IL
Sbjct: 238 PFVESPAAA--VASPRLKVSPDSLQYPPGYVGAVPDRSR--GSDEDDCGVSAMGYLTSIL 405
Query: 115 SAKVYDVAIESPLQLAPKLSERIGVNLWLKREDLQPVFSFKLRGAYNMMVKLPKEL 170
++KVYDVA+ESPL+LAPKLSER+GVN+WLKREDLQPVFSFK+RGAYNMM KLPK++
Sbjct: 406 TSKVYDVAVESPLELAPKLSERLGVNVWLKREDLQPVFSFKIRGAYNMMAKLPKDV 573
>CA919056 similar to PIR|T04211|T04 probable threonine dehydratase (EC
4.2.1.16) T5C23.70 [similarity] - Arabidopsis thaliana,
partial (47%)
Length = 799
Score = 70.5 bits (171), Expect = 2e-12
Identities = 43/134 (32%), Positives = 76/134 (56%), Gaps = 12/134 (8%)
Frame = -1
Query: 283 GGGLIAGIAAYVKTVSPQVKIIGVEPTDANTMALSLHHDQRVILDQVGGFADGVAVKEVG 342
GGGLI+G+A K+++P ++I+ EP A+ A S + + L + ADG+ +G
Sbjct: 502 GGGLISGVALAAKSINPAIRILAAEPKGADDAAQSKAAGRIITLPETNTIADGLRA-FLG 326
Query: 343 EETFRLCKDLIDGVVLVNRDSICASIKDMFEENRSILEPSGALALAG--AEAY------- 393
+ T+ + +DL+D ++ V I +++ +E + ++EPSGA+ LA +E++
Sbjct: 325 DFTWPVVRDLVDDIITVEDSEIVKAMQLCYEILKIVVEPSGAIGLAAVLSESFQKNPAWK 146
Query: 394 -CKYYGI--KGGNV 404
CK+ GI GGNV
Sbjct: 145 DCKHIGIVVSGGNV 104
>TC84921 similar to PIR|T04211|T04211 probable threonine dehydratase (EC
4.2.1.16) T5C23.70 [similarity] - Arabidopsis thaliana,
partial (32%)
Length = 696
Score = 68.6 bits (166), Expect = 7e-12
Identities = 43/140 (30%), Positives = 69/140 (48%)
Frame = +2
Query: 112 NILSAKVYDVAIESPLQLAPKLSERIGVNLWLKREDLQPVFSFKLRGAYNMMVKLPKELL 171
N A++ + +++P+ + L+ G L+ K E Q +FK RGA N + L E
Sbjct: 104 NEAHARIKSLILKTPVLSSTSLNAISGRQLYFKCESFQKGGAFKFRGACNAVFSLNDEDA 283
Query: 172 EKGVICSSAGNHAQGVALAAKRLNCNAVVAMPVTTPDIKWKSVEKLGATVVLIGDSYDEA 231
KGVI S+GNHA +ALAAK A + +P P K ++V++ G V+ +
Sbjct: 284 SKGVITHSSGNHAAALALAAKLRGIPAYIVIPKNAPTCKIENVKRYGGQVIWSEANMRSR 463
Query: 232 QAYAKKRGIEEGRTFVPAFD 251
+ A K E G F+ ++
Sbjct: 464 EETANKVWQETGAIFIHPYN 523
>TC86072 similar to GP|11559260|dbj|BAB18760. beta-cyanoalanine synthase
{Solanum tuberosum}, partial (96%)
Length = 1636
Score = 65.1 bits (157), Expect = 8e-11
Identities = 69/282 (24%), Positives = 119/282 (41%), Gaps = 10/282 (3%)
Frame = +3
Query: 43 FIAATISKPAEITPSPSSSSAAVGDH--LSSPRLRVSPDSLQYEPGYLGAVPDRSRSDGG 100
F+ A++ + T S SSSS AV + RL + + + + D + G
Sbjct: 78 FLMASLMSFLKRTSSSSSSSLAVACQHPMMMRRLMTTTATSADSSSFAQRIRDLPKDLPG 257
Query: 101 SDSDAMLTAMANILSAKVYDVAIESPLQLAPKLSERIGVNLWLKREDLQPVFSFKLRGAY 160
++ + V + +PL K++E G + +K+E +QP S K R A
Sbjct: 258 TN-----------IKKHVSQLIGRTPLVYLNKVTEGCGAYIAVKQEMMQPTASIKDRPAL 404
Query: 161 NMMVKLPKE-LLEKG---VICSSAGNHAQGVALAAKRLNCNAVVAMPVTTPDIKWKSVEK 216
MM K+ L+ G +I ++GN +A A V+ MP T + +
Sbjct: 405 AMMEDAEKKNLITPGKTILIEPTSGNMGISLAFMAAMKGYKMVLTMPSYTSLERRVCMRA 584
Query: 217 LGATVVLIGDSYDEAQAYAKKRGIEEGRT---FVPAFDHP-DVIMGQGTVGMEIMRQMVD 272
GA ++L + K + E + F +P + + T G EI
Sbjct: 585 FGAELILTDPTKGMGGTVKKAYDLLESTPNAFMLQQFSNPANTKVHFETTGPEIWEDTNG 764
Query: 273 PVHAIFVPVGGGGLIAGIAAYVKTVSPQVKIIGVEPTDANTM 314
V + +G GG ++G+ Y+K+ +P VKI GVEP+++N +
Sbjct: 765 QVDIFVMGIGSGGTVSGVGQYLKSQNPNVKIYGVEPSESNVL 890
>TC91866 similar to SP|O22682|CYSN_ARATH Probable cysteine synthase
chloroplast precursor (EC 4.2.99.8) (O- acetylserine
sulfhydrylase), partial (64%)
Length = 997
Score = 56.2 bits (134), Expect = 3e-08
Identities = 49/199 (24%), Positives = 83/199 (41%), Gaps = 8/199 (4%)
Frame = +3
Query: 125 SPLQLAPKLSERIGVNLWLKREDLQPVFSFKLRGAYNMMVKLPKE-LLEKG---VICSSA 180
+P+ K+++ N+ K E ++P S K R Y+M+ + + G ++ +
Sbjct: 213 TPMIYLNKVTDGCVANVAAKLESMEPCRSVKDRIGYSMLADAEESGAISPGKTVLVEPTT 392
Query: 181 GNHAQGVALAAKRLNCNAVVAMPVTTPDIKWKSVEKLGATVVLIGDSYDEAQAYAKKRGI 240
GN G+A A +V MP + + + GA VVL A K I
Sbjct: 393 GNTGLGLAFVAATKGYKLIVTMPASVNIERRILLRTFGAEVVLTNAEKGLKGAVDKTEEI 572
Query: 241 EEGRTFVPAFDHPD----VIMGQGTVGMEIMRQMVDPVHAIFVPVGGGGLIAGIAAYVKT 296
G F D + T G EI + V + +G GG I G Y+K
Sbjct: 573 VYGTPNAYMFRQFDNRNNTKIHFETTGPEIWEDTMGNVDLLVAAIGTGGTITGTGQYLKL 752
Query: 297 VSPQVKIIGVEPTDANTMA 315
++ ++K++GVEP D + ++
Sbjct: 753 MNKKIKVVGVEPADRSIIS 809
>TC85828 homologue to GP|18252506|gb|AAL66291.1 cysteine synthase {Glycine
max}, complete
Length = 1202
Score = 52.4 bits (124), Expect = 5e-07
Identities = 68/285 (23%), Positives = 121/285 (41%), Gaps = 10/285 (3%)
Frame = +3
Query: 114 LSAKVYDVAIESPLQLAPKLSERIGVNLWLKREDLQPVFSFKLRGAYNMMVKLP-KELLE 172
++ V ++ ++PL +L++ + K E ++P S K R Y+M+ K L+
Sbjct: 120 IAKDVTELIGKTPLVYLNRLADGCVARVAAKLELMEPCSSVKDRIGYSMIADAEEKGLIT 299
Query: 173 KG---VICSSAGNHAQGVALAAKRLNCNAVVAMPVTTPDIKWKSVEKLGATVVLIGDSYD 229
G +I ++GN G+A A ++ MP + + + GA +VL +
Sbjct: 300 PGQSVLIEPTSGNTGIGLAFMAAAKGYKLIITMPASMSLERRIILLAFGAELVLTDPAKG 479
Query: 230 EAQAYAKKRGI---EEGRTFVPAFDHP-DVIMGQGTVGMEIMRQMVDPVHAIFVPVGGGG 285
A K + + F++P + + T G EI + + A +G GG
Sbjct: 480 MKGAVQKAEELLAKTPNAYILQQFENPANPKVHYETTGPEIWKGTDGKIDAFVSGIGTGG 659
Query: 286 LIAGIAAYVKTVSPQVKIIGVEPTDANTMALSLHHDQRVILDQVG-GFADGVAVKEVGEE 344
I G Y+K + +K+IGVEP ++ ++ ++ +G GF GV
Sbjct: 660 TITGAGKYLKEQNSNIKLIGVEPVESPVLSGGKPGPHKI--QGIGAGFVPGV-------- 809
Query: 345 TFRLCKDLIDGVVLVNRDSICASIKDM-FEENRSILEPSGALALA 388
L +LID VV ++ D + K + +E + SGA A A
Sbjct: 810 ---LEVNLIDEVVQISSDEAIETAKLLALKEGLFVGISSGAAAAA 935
>TC88533 similar to GP|12081919|dbj|BAB20862. plastidic cysteine synthase 1
{Solanum tuberosum}, partial (65%)
Length = 1136
Score = 47.4 bits (111), Expect = 2e-05
Identities = 48/218 (22%), Positives = 92/218 (42%), Gaps = 5/218 (2%)
Frame = +1
Query: 179 SAGNHAQGVALAAKRLNCNAVVAMPVTTPDIKWKSVEKLGATVVLIGDSYDEAQAYAKKR 238
++GN G+A A ++ MP + + ++ GA +VL + A K
Sbjct: 37 TSGNTGIGLAFIAASKGYKLILTMPASMSLERRVLLKAFGAELVLTEAAKGMNGAVQKAE 216
Query: 239 GIEEGRT---FVPAFDHP-DVIMGQGTVGMEIMRQMVDPVHAIFVPVGGGGLIAGIAAYV 294
I + + FD+P + + T G EI + + +G GG I+G ++
Sbjct: 217 EIVKSTPDAYMLQQFDNPSNPKIHFETTGPEIWEDTRGKIDILVAGIGTGGTISGTGRFL 396
Query: 295 KTVSPQVKIIGVEPTDANTMALSLHHDQRVILDQVGGFADGVAVKEVGEETFRLCKDLID 354
K + +V++IGVEP ++N ++ + + G G + + EE ++D
Sbjct: 397 KQQNSKVQVIGVEPLESNILSGGKPGPHK-----IQGIGAGFVPRNLDEE-------VLD 540
Query: 355 GVVLVNRDSICASIKDM-FEENRSILEPSGALALAGAE 391
V+ ++ D + K + +E + SGA A A +
Sbjct: 541 EVIAISSDEAIETTKQIALQEGLLVGISSGAAAAAALQ 654
>TC76602 similar to SP|Q43317|CYSK_CITLA Cysteine synthase (EC 4.2.99.8)
(O-acetylserine sulfhydrylase) (O-acetylserine
(Thiol)-lyase), partial (97%)
Length = 1355
Score = 44.7 bits (104), Expect = 1e-04
Identities = 49/197 (24%), Positives = 80/197 (39%), Gaps = 10/197 (5%)
Frame = +2
Query: 125 SPLQLAPKLSERIGVNLWLKREDLQPVFSFKLRGAYNMMVKLP-KELLEKG---VICSSA 180
+PL ++E + K E LQ S K R + +M+ K L+ G ++ ++
Sbjct: 233 TPLVYLNNITEGCVARIAAKLEYLQSCCSVKDRISLSMIEDAESKGLITPGKTVLVEPTS 412
Query: 181 GNHAQGVALAAKRLNCNAVVAMPVTTPDIKWKSVEKLGATVVL------IGDSYDEAQAY 234
GN G+A A +V MP T + + GA V L I + +A
Sbjct: 413 GNTGIGLASIAAMRGYKLLVTMPATMSLERKIILRAFGAEVYLTDPAKGIDGVFQKADEL 592
Query: 235 AKKRGIEEGRTFVPAFDHPDVIMGQGTVGMEIMRQMVDPVHAIFVPVGGGGLIAGIAAYV 294
K +P + T G EI + V A+ +G GG + G ++
Sbjct: 593 LAKTPNSYKLNQFENSANPKI--HYETTGPEIWKDSGGRVDALVAGIGTGGTVTGTGKFL 766
Query: 295 KTVSPQVKIIGVEPTDA 311
K +P +K+ GVEPT++
Sbjct: 767 KEKNPDIKVYGVEPTES 817
>TC82671 similar to GP|8778327|gb|AAF79336.1| F14J16.13 {Arabidopsis
thaliana}, partial (33%)
Length = 685
Score = 37.0 bits (84), Expect = 0.022
Identities = 36/111 (32%), Positives = 48/111 (42%), Gaps = 7/111 (6%)
Frame = +1
Query: 118 VYDVAIESPLQLAPKLSERIGVNLWLKREDLQPVFSFKLRGAYNMMVKLPKELLEKG--- 174
+ D +PL LS+ G + K E L P S K R A V++ +E LE G
Sbjct: 127 IIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVA----VQIIEEALESGQLR 294
Query: 175 ----VICSSAGNHAQGVALAAKRLNCNAVVAMPVTTPDIKWKSVEKLGATV 221
V SAG+ A +A A C V +P K + +E LGATV
Sbjct: 295 RGGIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATV 447
>BQ144314 weakly similar to GP|21322711|em pherophorin-dz1 protein {Volvox
carteri f. nagariensis}, partial (6%)
Length = 1154
Score = 32.7 bits (73), Expect = 0.41
Identities = 20/53 (37%), Positives = 27/53 (50%), Gaps = 2/53 (3%)
Frame = +1
Query: 10 THPHHPSPPPLLRRHAFHKMPTNVLHPNPRRK--PFIAATISKPAEITPSPSS 60
+HP P PPP L H + P HPN ++K PF + S P I+ PS+
Sbjct: 19 SHPSSPPPPPFLPPHHPNCQPPP--HPNNKKKPPPFPPFSFSTPFPISFLPSN 171
>BQ141733 homologue to GP|20161357|dbj putative RNA helicase {Oryza sativa
(japonica cultivar-group)}, partial (3%)
Length = 1231
Score = 31.6 bits (70), Expect = 0.92
Identities = 21/58 (36%), Positives = 27/58 (46%), Gaps = 1/58 (1%)
Frame = -2
Query: 10 THPHHPSPPPLLRRHAFHKMPTNVLH-PNPRRKPFIAATISKPAEITPSPSSSSAAVG 66
THP HP L H F +PT +L P R +P AA + PA + S + VG
Sbjct: 192 THPSHPGTDNL---HKFKLVPTAILQSPGRRIRPLFAADPAAPAVRSRPMSRHAVEVG 28
>BF635948 similar to GP|21622312|emb putative protein {Neurospora crassa},
partial (6%)
Length = 538
Score = 31.6 bits (70), Expect = 0.92
Identities = 19/53 (35%), Positives = 24/53 (44%), Gaps = 1/53 (1%)
Frame = +1
Query: 12 PHHPSPPPL-LRRHAFHKMPTNVLHPNPRRKPFIAATISKPAEITPSPSSSSA 63
P+ P P P H F P +PNP P I++ A P PSSSS+
Sbjct: 112 PNQPPPSPTPFDMHTFFNPP----NPNPNPNPNISSQFPSSAPSPPPPSSSSS 258
>TC82241 similar to PIR|B84620|B84620 hypothetical protein At2g23070
[imported] - Arabidopsis thaliana, partial (79%)
Length = 1468
Score = 31.2 bits (69), Expect = 1.2
Identities = 19/57 (33%), Positives = 27/57 (47%)
Frame = +3
Query: 20 LLRRHAFHKMPTNVLHPNPRRKPFIAATISKPAEITPSPSSSSAAVGDHLSSPRLRV 76
LLR H H++ T + P P + F+ T +P P SSSAA H S ++
Sbjct: 72 LLRHHHHHQLSTTLFRP-PLLRNFVLTT-------SPPPISSSAAAARHSGSTNRKI 218
>AW697223 similar to GP|9757890|dbj phosphoribosylanthranilate
transferase-like protein {Arabidopsis thaliana}, partial
(5%)
Length = 778
Score = 31.2 bits (69), Expect = 1.2
Identities = 24/86 (27%), Positives = 36/86 (40%)
Frame = -1
Query: 2 AALRFSTTTHPHHPSPPPLLRRHAFHKMPTNVLHPNPRRKPFIAATISKPAEITPSPSSS 61
A+L THPHHP P R P+ L P P + + P ++ SSS
Sbjct: 523 ASLAVPPPTHPHHPPRPSTRRLRVPTSSPSYSLPPPPHHSFLLPR--AGPCFMS**TSSS 350
Query: 62 SAAVGDHLSSPRLRVSPDSLQYEPGY 87
+ L SP L D++ ++ G+
Sbjct: 349 QTSPKISLRSPTLN---DNVSFQTGF 281
>BQ144079 similar to GP|6523547|emb| hydroxyproline-rich glycoprotein DZ-HRGP
{Volvox carteri f. nagariensis}, partial (12%)
Length = 1071
Score = 31.2 bits (69), Expect = 1.2
Identities = 19/77 (24%), Positives = 28/77 (35%)
Frame = +1
Query: 11 HPHHPSPPPLLRRHAFHKMPTNVLHPNPRRKPFIAATISKPAEITPSPSSSSAAVGDHLS 70
HP+HP P R + P P P P +A S +P P + + L
Sbjct: 568 HPNHPHPNARKRTQTLSRPP-----PPPTPPPPVAQPKSGTPPPSPYPKKPNPILSQSLR 732
Query: 71 SPRLRVSPDSLQYEPGY 87
+P + + P P Y
Sbjct: 733 TPPIAIPPPPAPQTPQY 783
>TC89433 similar to GP|13277220|emb|CAC34409. ZF-HD homeobox protein
{Flaveria bidentis}, partial (52%)
Length = 1430
Score = 30.8 bits (68), Expect = 1.6
Identities = 19/62 (30%), Positives = 27/62 (42%)
Frame = +1
Query: 11 HPHHPSPPPLLRRHAFHKMPTNVLHPNPRRKPFIAATISKPAEITPSPSSSSAAVGDHLS 70
H HHP PPPL + + + P P + +S P ++AA +HLS
Sbjct: 703 HRHHPPPPPLFQSRSPSSPSPPPISSYP-SAPHMLLALSGAGVGLSIPPENTAAPLNHLS 879
Query: 71 SP 72
SP
Sbjct: 880 SP 885
>TC76916 MtN4
Length = 1850
Score = 30.8 bits (68), Expect = 1.6
Identities = 23/80 (28%), Positives = 30/80 (36%), Gaps = 1/80 (1%)
Frame = +1
Query: 7 STTTHPHHPSPPPLLRRHAFHKMPTNVLHPNPRRKPFI-AATISKPAEITPSPSSSSAAV 65
ST PHHP PP + H P V P + P + + KP + P P V
Sbjct: 658 STPKPPHHPKPPAVHPPHVPKPHPPYVPKPPIVKPPIVHPPYVPKPPVVKPPPYVPKPPV 837
Query: 66 GDHLSSPRLRVSPDSLQYEP 85
P+ V P + Y P
Sbjct: 838 VRPPYVPKPPVVPVTPPYVP 897
>TC86141 similar to GP|20260580|gb|AAM13188.1 unknown protein {Arabidopsis
thaliana}, partial (82%)
Length = 1876
Score = 30.4 bits (67), Expect = 2.0
Identities = 19/53 (35%), Positives = 26/53 (48%), Gaps = 4/53 (7%)
Frame = +1
Query: 12 PHHPSPPPLLRR---HAFH-KMPTNVLHPNPRRKPFIAATISKPAEITPSPSS 60
PH+P PPP + H +PT P+ ++ F KP+ IT SPSS
Sbjct: 295 PHYPPPPPYKSTPTPSSLHFSIPTTRNLPSSLKRHFSFKN*KKPSVITTSPSS 453
>BQ144407 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (23%)
Length = 1217
Score = 30.4 bits (67), Expect = 2.0
Identities = 17/56 (30%), Positives = 24/56 (42%)
Frame = -3
Query: 5 RFSTTTHPHHPSPPPLLRRHAFHKMPTNVLHPNPRRKPFIAATISKPAEITPSPSS 60
R S H SPPP H + P P I A ++P+ +TP+P+S
Sbjct: 726 RLSRCVSHLHSSPPPPTLIHPLPPPSPYPISPPPSLDISITAPSTRPSSLTPTPAS 559
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,153,844
Number of Sequences: 36976
Number of extensions: 241217
Number of successful extensions: 1858
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 1765
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1835
length of query: 606
length of database: 9,014,727
effective HSP length: 102
effective length of query: 504
effective length of database: 5,243,175
effective search space: 2642560200
effective search space used: 2642560200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0375a.8


