

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
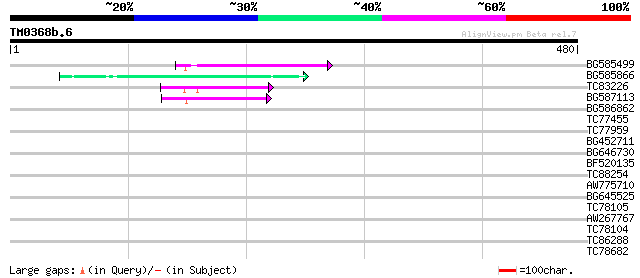
Query= TM0368b.6
(480 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG585499 70 2e-12
BG585866 62 5e-10
TC83226 weakly similar to PIR|G86419|G86419 probable reverse tra... 52 5e-07
BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At... 52 7e-07
BG586862 39 0.006
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 37 0.013
TC77959 similar to GP|1621268|emb|CAB02653.1 unknown {Ricinus co... 34 0.14
BG452711 33 0.24
BG646730 30 2.7
BF520135 29 4.6
TC88254 weakly similar to GP|9294409|dbj|BAB02490.1 polygalactur... 29 4.6
AW775710 similar to PIR|T46096|T46 hypothetical protein T25B15.3... 28 6.0
BG645525 homologue to GP|19568098|gb osmotic stress-activated pr... 28 7.8
TC78105 homologue to GP|19568098|gb|AAL89456.1 osmotic stress-ac... 28 7.8
AW267767 weakly similar to GP|20465617|gb| unknown protein {Arab... 28 7.8
TC78104 homologue to GP|19568098|gb|AAL89456.1 osmotic stress-ac... 28 7.8
TC86288 similar to GP|2407790|gb|AAB70660.1| grr1 {Glycine max},... 28 7.8
TC78682 similar to GP|13898655|gb|AAK48848.1 expansin {Prunus ce... 28 7.8
>BG585499
Length = 792
Score = 70.1 bits (170), Expect = 2e-12
Identities = 38/136 (27%), Positives = 63/136 (45%), Gaps = 3/136 (2%)
Frame = +3
Query: 141 SAYELLT--ENIEDTGPDPMWCLLWRWKGAPCVKMFLWKVLNNGLMVNVKRVHNHLADSD 198
S+Y LL ++I D W +LW W+G + F+W V + ++ N +R
Sbjct: 177 SSYNLLV*DQSIVDCD----WKMLWGWRGPHRTQTFMWLVAHGCILTNYRRSRWGTRVLA 344
Query: 199 LCPLCGTLSESVIHALRDCPMVKPVRLANLPTGGESHFFSM-ELQAWLLRNMRDGLQGWD 257
CP CG E+V+H L DC V + +P+ ++FFS + + W+ +N+ G
Sbjct: 345 TCPCCGNADETVLHVLCDCRPASQVWIRLVPSDWITNFFSFDDCRDWVFKNLSKRSNGVS 524
Query: 258 RDAWTKRFAINVWCLW 273
+ W F W +W
Sbjct: 525 KFKWQPTFMTTCWHMW 572
>BG585866
Length = 828
Score = 62.0 bits (149), Expect = 5e-10
Identities = 51/212 (24%), Positives = 82/212 (38%), Gaps = 1/212 (0%)
Frame = +3
Query: 43 GIRWQIGDGKVVRF*QDRWLASGILLAEVAVQPIPPGSLNRVVDEFLGSNGTWETAGFVE 102
G W+ G G F W + G+L + I L V D F + G T
Sbjct: 114 GYTWRAGSGNS-SFWYTNWSSLGLLGTQAPFVDIHDLHLT-VKDVF--TTGGQHTQSLYT 281
Query: 103 LMPNIVFQEIMESLPRHLSSAPGLPVWGRTTSGINSTASAYELLTENIEDTG-PDPMWCL 161
++P + + I + +S +W ++G+ + S Y + E + W
Sbjct: 282 ILPTDIAEVINNTHLNFNASIGDAYIWPHNSNGVYTAKSGYSWILSQTETVNYNNSSWSW 461
Query: 162 LWRWKGAPCVKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVK 221
+WR K K FLW +N + H ++ +S +C CG ES H +RDC K
Sbjct: 462 IWRLKIPEKYKFFLWLACHNAVPTLSLLNHRNMVNSAICSRCGEHEESFFHCVRDCRFSK 641
Query: 222 PVRLANLPTGGESHFFSMELQAWLLRNMRDGL 253
+ + F S +Q WL +DG+
Sbjct: 642 -IIWHKIGFSSPDFFSSSSVQDWL----KDGI 722
>TC83226 weakly similar to PIR|G86419|G86419 probable reverse transcriptase
100033-105622 [imported] - Arabidopsis thaliana, partial
(2%)
Length = 885
Score = 52.0 bits (123), Expect = 5e-07
Identities = 30/104 (28%), Positives = 46/104 (43%), Gaps = 8/104 (7%)
Frame = +3
Query: 128 VWGRTTSGINSTASAYELL----TENIEDTGPDP----MWCLLWRWKGAPCVKMFLWKVL 179
+W +GI S S Y L T+ I +T +W +W P K+ LW++L
Sbjct: 3 MWMHNPTGIYSVKSGYNTLRTWQTQQINNTSTSSDETLIWKKIWSLHTIPRHKVLLWRIL 182
Query: 180 NNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPV 223
N+ L V + LCP C + +E++ H CP+ K V
Sbjct: 183 NDSLPVRSSLRKRGIQCYPLCPRCHSKTETITHLFMSCPLSKRV 314
>BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (10%)
Length = 767
Score = 51.6 bits (122), Expect = 7e-07
Identities = 27/102 (26%), Positives = 45/102 (43%), Gaps = 9/102 (8%)
Frame = -3
Query: 129 WGRTTSGINSTASAYELLTE---------NIEDTGPDPMWCLLWRWKGAPCVKMFLWKVL 179
W + SG S S Y + T ++ D ++ +W++ +P V+ FLW+ +
Sbjct: 414 WEYSKSGHYSVKSGYYVQTNIIAAANQRGTVDQPSLDDLYQRVWKYNTSPKVRHFLWRCI 235
Query: 180 NNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVK 221
+N L H++ C CG SE+V H L CP +
Sbjct: 234 SNSLPTAANMRSRHISKDGSCSRCGMESETVNHILFQCPYAR 109
>BG586862
Length = 804
Score = 38.5 bits (88), Expect = 0.006
Identities = 20/60 (33%), Positives = 31/60 (51%)
Frame = -1
Query: 162 LWRWKGAPCVKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVK 221
+W K P K FLW++L+N L V + + S LCP C + E+V H +C + +
Sbjct: 654 VWGIKTIPRHKSFLWRLLHNALPVKDELHKRGIRCSLLCPRCESKIETVQHLFLNCEVTQ 475
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 37.4 bits (85), Expect = 0.013
Identities = 31/104 (29%), Positives = 46/104 (43%), Gaps = 8/104 (7%)
Frame = -2
Query: 128 VWGRTTSGINSTASAYELLTEN--IEDT---GPDPMWCLLWRWKGAPCVKMFLWKVLNNG 182
VW G+ S S Y LL +ED + ++ LW+ K V F W + +
Sbjct: 439 VWKPDKEGVFSVNSCYFLLQNLRLLEDRLSYEEEVIFRELWKSKAPAKVLAFSWTLFLDR 260
Query: 183 L--MVNV-KRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPV 223
+ MVN+ KR + DS C CG E+V+H C ++ V
Sbjct: 259 IPTMVNLGKRRLLRVEDSKRCVFCGCQDETVVHLFLHCDVISKV 128
>TC77959 similar to GP|1621268|emb|CAB02653.1 unknown {Ricinus communis},
partial (87%)
Length = 1224
Score = 33.9 bits (76), Expect = 0.14
Identities = 31/98 (31%), Positives = 46/98 (46%), Gaps = 3/98 (3%)
Frame = +1
Query: 35 NTWESFSCGIRWQIGDGKVV--RF*QDRWLASGILLAEVAVQPIPPGSLNRVVDEFLGSN 92
N WESF+CG R +G++V + A G L+ VA++ + P R L N
Sbjct: 481 NGWESFACGFRADYPEGEIVIHNSGVEDDPACGPLIDSVALKVLNPPIRTRA---NLLKN 651
Query: 93 GTWETAGFVELMPNIVFQEIMESLPRHLSSAPG-LPVW 129
G +E +V PN + + +P H+ A G LP W
Sbjct: 652 GNFEEGPYV--FPNASWGVL---IPPHIEDAHGPLPGW 750
>BG452711
Length = 672
Score = 33.1 bits (74), Expect = 0.24
Identities = 13/31 (41%), Positives = 19/31 (60%)
Frame = +3
Query: 193 HLADSDLCPLCGTLSESVIHALRDCPMVKPV 223
+L S+LCP+C S+ ++HAL C K V
Sbjct: 387 NLISSNLCPICNQRSQDMLHALFSCTRAKDV 479
>BG646730
Length = 799
Score = 29.6 bits (65), Expect = 2.7
Identities = 13/36 (36%), Positives = 18/36 (49%)
Frame = +1
Query: 27 SSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDRWL 62
S W + N + G RW I +G+ VR +D WL
Sbjct: 400 SYAWRSMFNIKDVIDLGSRWSISNGQNVRIWKDDWL 507
>BF520135
Length = 202
Score = 28.9 bits (63), Expect = 4.6
Identities = 21/68 (30%), Positives = 33/68 (47%), Gaps = 6/68 (8%)
Frame = +3
Query: 159 WCLL-WRWKGAPCVKMFLWKVLNNGL--MVNVKR---VHNHLADSDLCPLCGTLSESVIH 212
WCLL WR + V +F W++ + L NV + VH+ D+ +C L ES +H
Sbjct: 3 WCLLLWRKEDLSKVSIFAWRLFHGRLPTKANVFKRGIVHH---DAHMCVTRCRLIESDVH 173
Query: 213 ALRDCPMV 220
C ++
Sbjct: 174 LFLHCDVL 197
>TC88254 weakly similar to GP|9294409|dbj|BAB02490.1 polygalacturonase
inhibitor-like protein {Arabidopsis thaliana}, partial
(54%)
Length = 706
Score = 28.9 bits (63), Expect = 4.6
Identities = 11/38 (28%), Positives = 23/38 (59%), Gaps = 1/38 (2%)
Frame = +3
Query: 10 YGCG-NNVMSIVSHRASESSIWNGVRNTWESFSCGIRW 46
+GC ++ +++S +AS ++G+ NTW +C + W
Sbjct: 60 HGCSPSDRTALLSFKASLKEPYHGIFNTWSGENCCVNW 173
>AW775710 similar to PIR|T46096|T46 hypothetical protein T25B15.30 -
Arabidopsis thaliana, partial (29%)
Length = 492
Score = 28.5 bits (62), Expect = 6.0
Identities = 12/25 (48%), Positives = 16/25 (64%)
Frame = -2
Query: 346 LDRRLLSSSGYFKPLTCGAMGNSHG 370
L R ++SSS Y + CGA G+ HG
Sbjct: 341 LFRAIMSSSSYKTSMVCGASGSFHG 267
>BG645525 homologue to GP|19568098|gb osmotic stress-activated protein kinase
{Nicotiana tabacum}, partial (63%)
Length = 811
Score = 28.1 bits (61), Expect = 7.8
Identities = 11/30 (36%), Positives = 19/30 (62%)
Frame = +3
Query: 253 LQGWDRDAWTKRFAINVWCLWRMM*YSIMC 282
LQG D++T ++ V C WR +* +++C
Sbjct: 312 LQGGGFDSYTSWYSYGVCCWWRAL*PNLLC 401
>TC78105 homologue to GP|19568098|gb|AAL89456.1 osmotic stress-activated
protein kinase {Nicotiana tabacum}, partial (35%)
Length = 731
Score = 28.1 bits (61), Expect = 7.8
Identities = 11/30 (36%), Positives = 19/30 (62%)
Frame = +3
Query: 253 LQGWDRDAWTKRFAINVWCLWRMM*YSIMC 282
LQG D++T ++ V C WR +* +++C
Sbjct: 498 LQGGGFDSYTSWYSYGVCCWWRAL*PNLLC 587
>AW267767 weakly similar to GP|20465617|gb| unknown protein {Arabidopsis
thaliana}, partial (5%)
Length = 737
Score = 28.1 bits (61), Expect = 7.8
Identities = 18/53 (33%), Positives = 27/53 (49%), Gaps = 2/53 (3%)
Frame = +2
Query: 258 RDAWTK--RFAINVWCLWRMM*YSIMCSGIVLRYNAGLCRASMMMVWLPLMLS 308
+ AW R + N++ LWR M S +CS I++ RA M+ P +LS
Sbjct: 131 KGAWDSCFRLSTNIYLLWRRMLKSTICS*III-IEIPFLRARMVERTTPSLLS 286
>TC78104 homologue to GP|19568098|gb|AAL89456.1 osmotic stress-activated
protein kinase {Nicotiana tabacum}, partial (95%)
Length = 1434
Score = 28.1 bits (61), Expect = 7.8
Identities = 11/30 (36%), Positives = 19/30 (62%)
Frame = +2
Query: 253 LQGWDRDAWTKRFAINVWCLWRMM*YSIMC 282
LQG D++T ++ V C WR +* +++C
Sbjct: 338 LQGGGFDSYTSWYSYGVCCWWRAL*PNLLC 427
>TC86288 similar to GP|2407790|gb|AAB70660.1| grr1 {Glycine max}, partial
(84%)
Length = 2900
Score = 28.1 bits (61), Expect = 7.8
Identities = 16/69 (23%), Positives = 35/69 (50%), Gaps = 3/69 (4%)
Frame = +1
Query: 167 GAPCVKMF-LWKV--LNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPV 223
G P +K F LW V +++ ++ + + + + DLC L +++I + CP + +
Sbjct: 742 GCPSLKSFTLWDVATISDAGLIEIANGCHQIENLDLCKLPTISDKALIAVAKHCPNLTEL 921
Query: 224 RLANLPTGG 232
+ + P+ G
Sbjct: 922 SIESCPSIG 948
>TC78682 similar to GP|13898655|gb|AAK48848.1 expansin {Prunus cerasus},
partial (91%)
Length = 1236
Score = 28.1 bits (61), Expect = 7.8
Identities = 11/18 (61%), Positives = 11/18 (61%)
Frame = +3
Query: 159 WCLLWRWKGAPCVKMFLW 176
WCLLW W A C FLW
Sbjct: 168 WCLLW-WSMARCSCYFLW 218
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.334 0.143 0.505
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,727,557
Number of Sequences: 36976
Number of extensions: 302815
Number of successful extensions: 2394
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 2351
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2391
length of query: 480
length of database: 9,014,727
effective HSP length: 100
effective length of query: 380
effective length of database: 5,317,127
effective search space: 2020508260
effective search space used: 2020508260
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0368b.6


