

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0367.1
(481 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
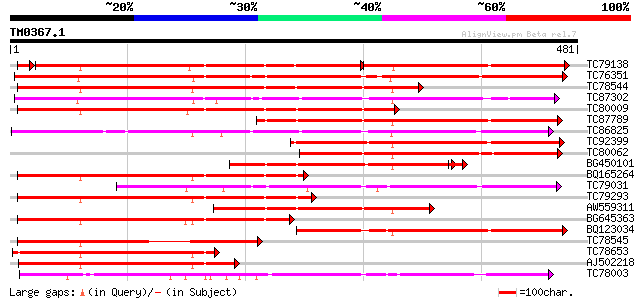
Score E
Sequences producing significant alignments: (bits) Value
TC79138 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:... 314 e-146
TC76351 similar to GP|7635494|emb|CAB88666.1 putative UDP-glycos... 401 e-112
TC78544 weakly similar to PIR|T03747|T03747 glucosyltransferase ... 389 e-108
TC87302 similar to GP|19911199|dbj|BAB86926. glucosyltransferase... 362 e-100
TC80009 weakly similar to PIR|T03747|T03747 glucosyltransferase ... 333 1e-91
TC87789 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:... 328 2e-90
TC86825 similar to GP|18151384|dbj|BAB83692. ABA-glucosyltransfe... 305 2e-83
TC92399 weakly similar to GP|5763524|dbj|BAA83484.1 UDP-glucose:... 291 5e-79
TC80062 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:... 272 2e-73
BG450101 similar to GP|4115534|dbj UDP-glycose:flavonoid glycosy... 234 6e-64
BQ165264 weakly similar to PIR|T07786|T077 UDP-glucose glucosylt... 238 3e-63
TC79031 weakly similar to GP|13492674|gb|AAK28303.1 phenylpropan... 230 8e-61
TC79293 weakly similar to GP|15290056|dbj|BAB63750. putative glu... 222 2e-58
AW559311 weakly similar to PIR|T03747|T037 glucosyltransferase I... 213 1e-55
BG645363 weakly similar to GP|15290080|dbj putative salicylate-i... 209 2e-54
BQ123034 weakly similar to GP|14334982|gb putative glucosyltrans... 186 1e-47
TC78545 weakly similar to GP|13492676|gb|AAK28304.1 phenylpropan... 178 4e-45
TC78653 similar to PIR|T46162|T46162 glucosyltransferase-like pr... 177 8e-45
AJ502218 similar to GP|22208467|gb| putative glucosyl transferas... 177 1e-44
TC78003 similar to GP|20146093|dbj|BAB88935. glucosyltransferase... 168 5e-42
>TC79138 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:flavonoid
glycosyltransferase {Vigna mungo}, partial (36%)
Length = 1575
Score = 314 bits (804), Expect(3) = e-146
Identities = 159/285 (55%), Positives = 205/285 (71%), Gaps = 5/285 (1%)
Frame = +2
Query: 23 MIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPN---LHLHTVPFPSRQVGLPDGIES 79
M PLCDIA LFAS G HVTIITTPSN I S+P+ L LHTVPFPS QVGLP G+E+
Sbjct: 53 MTPLCDIATLFASCGHHVTIITTPSNAQIILKSIPSHNHLRLHTVPFPSHQVGLPLGVEN 232
Query: 80 FSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYCFPWIDDLTTELHIPRLSFN 139
+ ++ + K+H LLR PI F+E DC+VAD+ F W+D+L LHIPRL+FN
Sbjct: 233 LAFVNNVDNSCKIHHATMLLRSPINHFVEQDSPDCIVADFMFLWVDELANRLHIPRLAFN 412
Query: 140 PSPLFALCAMRA--KRGSSSFLISGLPHGDIALNASPPEALTACVEPLLRKELKSHGVLI 197
LFA+CAM + R S +I GLPH I LNA PP+ALT +EPLL ELKS+G+++
Sbjct: 413 GFSLFAICAMESLKARDFESSIIQGLPH-CITLNAMPPKALTKFMEPLLETELKSYGLIV 589
Query: 198 NSFVELDGHEYVKHYERTTGGHKAWLLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLS 257
N+F ELDG EY++HYE+T GH+AW LGP+SL+ T +EKA+RG + SVV E LSWL+
Sbjct: 590 NNFTELDGEEYIEHYEKTI-GHRAWHLGPSSLICRTTQEKADRG-QTSVVDVHECLSWLN 763
Query: 258 TKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEK 302
+K+P+SVLY+CFGS+C F+N+QL+E+A IEAS +F+WV P K
Sbjct: 764 SKQPNSVLYICFGSLCHFTNKQLYEIASAIEASGHQFIWVSPRRK 898
Score = 218 bits (555), Expect(3) = e-146
Identities = 108/177 (61%), Positives = 137/177 (77%), Gaps = 2/177 (1%)
Frame = +1
Query: 301 EKRGKEEEEEEEKEKWLPEGFEEK--GMILRGWAPQLLILGHPSVGAFLTHCGSNSALEA 358
EK+GKE+E +E EKW+P+GFEE+ GMI+RGWAPQ++ILGHP++GAFLTHCG NS +EA
Sbjct: 889 EKKGKEDESNDENEKWMPKGFEERNIGMIIRGWAPQVVILGHPAIGAFLTHCGWNSTVEA 1068
Query: 359 VSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKA 418
VSAGVP+ITWPV +QFY EKL+T+VRG+GVEVG EEWS G ++++ ++ RD IEKA
Sbjct: 1069 VSAGVPMITWPVHDEQFYNEKLITQVRGIGVEVGVEEWSFIGFMKKKK--IVGRDIIEKA 1242
Query: 419 VRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHLKTLRDSNSVSS 475
+RRLM GG EA +IR+R E A AVQ GGSS++NL ALI LK R S+ S
Sbjct: 1243 LRRLMDGGIEAVEIRKRAQEYAIKAKRAVQEGGSSHKNLMALIDDLKRQRGHKSLDS 1413
Score = 25.8 bits (55), Expect(3) = e-146
Identities = 10/15 (66%), Positives = 12/15 (79%)
Frame = +3
Query: 7 QRPLKLYFIPYLAAG 21
+RPLK +FIPY A G
Sbjct: 6 ERPLKFHFIPYPAPG 50
>TC76351 similar to GP|7635494|emb|CAB88666.1 putative UDP-glycose {Cicer
arietinum}, partial (92%)
Length = 1645
Score = 401 bits (1030), Expect = e-112
Identities = 217/491 (44%), Positives = 303/491 (61%), Gaps = 22/491 (4%)
Frame = +2
Query: 5 SEQRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLP------- 57
+E +PLK+Y +P+ A GH+IPL ++A L AS+ QHVTIITTPSN D ++
Sbjct: 80 TESKPLKIYMLPFFAQGHLIPLVNLARLVASKNQHVTIITTPSNAQLFDKTIEEEKAAGH 259
Query: 58 NLHLHTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVA 117
++ +H + FPS Q+GLP G+E+ +++D QT K+H ++ I+ FM+ +P D ++
Sbjct: 260 HIRVHIIKFPSAQLGLPTGVENLFAASDNQTAGKIHMAAHFVKADIEEFMKENPPDVFIS 439
Query: 118 DYCFPWIDDLTTELHIPRLSFNPSPLFALCAMRAKRG--------SSSFLISGLPHGDIA 169
D F W + L IPRL FNP +F +C ++A + S + I GLPH +
Sbjct: 440 DIIFTWSESTAKNLQIPRLVFNPISIFDVCMIQAIQSHPESFVSDSGPYQIHGLPH-PLT 616
Query: 170 LNASPPEALTACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASL 229
L P E L+ E SHGV++NSF ELD Y ++YE TG K W +GP SL
Sbjct: 617 LPIKPSPGFARLTESLIEAENDSHGVIVNSFAELD-EGYTEYYENLTG-RKVWHVGPTSL 790
Query: 230 -VRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIE 288
V K+K S ++ + L+WL TK+PSSVLY+ FGS+C SNEQL EMA GIE
Sbjct: 791 MVEIPKKKKVVSTENDSSITKHQSLTWLDTKEPSSVLYISFGSLCRLSNEQLKEMANGIE 970
Query: 289 ASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGF------EEKGMILRGWAPQLLILGHPS 342
AS +F+WVV GKE E+E+ WLP+GF E+KGM+++GW PQ LIL HPS
Sbjct: 971 ASKHQFLWVV----HGKEGEDEDN---WLPKGFVERMKEEKKGMLIKGWVPQALILDHPS 1129
Query: 343 VGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGR 402
+G FLTHCG N+ +EA+S+GVP++T P GDQ+Y EKL+TEV +GVEVGA EWSM
Sbjct: 1130IGGFLTHCGWNATVEAISSGVPMVTMPGFGDQYYNEKLVTEVHRIGVEVGAAEWSM--SP 1303
Query: 403 RERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIA 462
+ ++ V+ ++IEKAV++LM E +IR+R E+ A AVQ GGSS LT L+
Sbjct: 1304YDAKKTVVRAERIEKAVKKLMDSNGEGGEIRKRAKEMKEKAWKAVQEGGSSQNCLTKLVD 1483
Query: 463 HLKTLRDSNSV 473
+L ++ + SV
Sbjct: 1484YLHSVVVTKSV 1516
>TC78544 weakly similar to PIR|T03747|T03747 glucosyltransferase IS5a (EC
2.4.1.-) salicylate-induced - common tobacco, partial
(26%)
Length = 1248
Score = 389 bits (998), Expect = e-108
Identities = 200/365 (54%), Positives = 261/365 (70%), Gaps = 20/365 (5%)
Frame = +3
Query: 7 QRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPN--LHLHTV 64
+RPLKL+FIPY A+GHM+PLCDIA LFASRGQHVTIITTPSN ++ +L + L LHTV
Sbjct: 90 ERPLKLHFIPYPASGHMMPLCDIATLFASRGQHVTIITTPSNAQSLTKTLSSAALRLHTV 269
Query: 65 PFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYCFPWI 124
FP +QV LP G+ES +S+ D TT+K+H G LL E + F+E +P DC++AD F W
Sbjct: 270 EFPYQQVDLPKGVESMTSTTDPITTWKIHNGAMLLNEAVGDFVEKNPPDCIIADSAFSWA 449
Query: 125 DDLTTELHIPRLSFNPSPLFALCAMRAKR-------------GSSSFLISGLPHGDIALN 171
+DL +L IP L+FN S LFA+ + R SSS+++ L H +I L
Sbjct: 450 NDLAHKLQIPNLTFNGSSLFAVSIFHSLRTNNLLHTDADADSDSSSYVVPNLHHDNITLC 629
Query: 172 ASPPEALTACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVR 231
+ PP+ L+ + +L LKS G +IN+FVELDG E VKHYE+TT GHKAW LGP S +R
Sbjct: 630 SKPPKVLSMFIGMMLDTVLKSTGYIINNFVELDGEECVKHYEKTT-GHKAWHLGPTSFIR 806
Query: 232 GTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASS 291
T +EKAE+G+ KS VS E L+WL +++ +SV+Y+CFGS+ FS++QL+E+AC +EAS
Sbjct: 807 KTVQEKAEKGN-KSDVSEHECLNWLKSQRVNSVVYICFGSINHFSDKQLYEIACAVEASG 983
Query: 292 VEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEE-----KGMILRGWAPQLLILGHPSVGAF 346
F+WVVP EK+GKE+E EEEKEKWLP+GFEE KG I+RGWAPQ+LIL +P+VG F
Sbjct: 984 HPFIWVVP-EKKGKEDEIEEEKEKWLPKGFEERNIGKKGFIIRGWAPQVLILSNPAVGGF 1160
Query: 347 LTHCG 351
LTHCG
Sbjct: 1161LTHCG 1175
Score = 29.6 bits (65), Expect = 2.7
Identities = 21/61 (34%), Positives = 26/61 (42%), Gaps = 1/61 (1%)
Frame = +2
Query: 314 EKWLPEGFEEKGMILRGWAPQLLILGHPSVGAFLTHCGSNSAL-EAVSAGVPIITWPVLG 372
EK EGF +G+ P + P G + L AV AGVP+ITWP
Sbjct: 1076 EKHWKEGFHNQGL-----GPTSSDIEQPCGGGISDTLRRETQL*RAVGAGVPMITWPCHA 1240
Query: 373 D 373
D
Sbjct: 1241 D 1243
>TC87302 similar to GP|19911199|dbj|BAB86926. glucosyltransferase-8 {Vigna
angularis}, partial (53%)
Length = 1787
Score = 362 bits (929), Expect = e-100
Identities = 210/489 (42%), Positives = 288/489 (57%), Gaps = 27/489 (5%)
Frame = +1
Query: 5 SEQRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSL--PNLHLH 62
+E + L + F P+LA GH+IP D+A +F+SRG VTI+TT N I ++ +++
Sbjct: 40 NENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKINIK 219
Query: 63 TVPFPS-RQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYCF 121
T+ FPS + GLP+G E+ S+ K + LLREP++ +E DC+VAD F
Sbjct: 220 TIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKPDCLVADMFF 399
Query: 122 PWIDDLTTELHIPRLSFNPSPLFALCAMRAKRG----------SSSFLISGLPHGDIALN 171
PW D + +IPR+ F+ F LC + R + F++ LP G+I L
Sbjct: 400 PWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLP-GEITLT 576
Query: 172 ASP-------PEALTACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLL 224
+ T +E E+KS GV+ N+F EL+ Y HY R G KAW L
Sbjct: 577 KMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEP-VYADHY-RNELGRKAWHL 750
Query: 225 GPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMA 284
GP SL +EKA RG E S+ E L WL +K+P+SV+YVCFGSM FS+ QL E+A
Sbjct: 751 GPVSLCNRDTEEKACRGREASI-DEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIA 927
Query: 285 CGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEE------KGMILRGWAPQLLIL 338
G+EAS V F+WVV K + E E +WLPEGFEE KG+I+RGWAPQ++IL
Sbjct: 928 MGLEASEVPFIWVVR-----KSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVMIL 1092
Query: 339 GHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEW-S 397
H SVG F+THCG NS LE VSAG+P++TWP+ G+QFY K ++++ +GV VG + W
Sbjct: 1093DHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIG 1272
Query: 398 MGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNL 457
MGGG + +D IEKAVRR+M G+EAE++R R E G +A AV+ GGSSY +
Sbjct: 1273MGGGE------PVKKDVIEKAVRRIM-VGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDF 1431
Query: 458 TALIAHLKT 466
+ LI LK+
Sbjct: 1432SNLIEDLKS 1458
>TC80009 weakly similar to PIR|T03747|T03747 glucosyltransferase IS5a (EC
2.4.1.-) salicylate-induced - common tobacco, partial
(19%)
Length = 1118
Score = 333 bits (853), Expect = 1e-91
Identities = 174/342 (50%), Positives = 232/342 (66%), Gaps = 18/342 (5%)
Frame = +3
Query: 7 QRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPN-----LHL 61
QRPLKL+FIP+LA+GHMIPL DIA +FAS G VT+ITTPSN ++ SL + L L
Sbjct: 96 QRPLKLHFIPFLASGHMIPLFDIATMFASHGHQVTVITTPSNAQSLTKSLSSAASFFLRL 275
Query: 62 HTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYCF 121
HTV FPS QV LP GIES SS+ D T++K+H+G LL PI+ FME P DC+++D +
Sbjct: 276 HTVDFPSEQVDLPKGIESMSSTTDSITSWKIHRGAMLLHGPIENFMEKDPPDCIISDSTY 455
Query: 122 PWIDDLTTELHIPRLSFNPSPLFALCAM-------------RAKRGSSSFLISGLPHGDI 168
PW +DL +L IP L+FN LF + + + SSSFL+ PH I
Sbjct: 456 PWANDLAHKLQIPNLTFNGLSLFTISLVESLIRNNLLHSDTNSDSDSSSFLVPNFPH-RI 632
Query: 169 ALNASPPEALTACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPAS 228
L+ PP+ L+ ++ +L LKS ++IN+F ELDG E ++HYE+TT G K W LGP S
Sbjct: 633 TLSEKPPKVLSKFLKMMLETVLKSKALIINNFAELDGEECIQHYEKTT-GRKVWHLGPTS 809
Query: 229 LVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIE 288
L+R T +EKAERG+E V+ E LSWL++++ ++VLY+CFGS+ S++QL+EMAC IE
Sbjct: 810 LIRRTIQEKAERGNE-GEVNMHECLSWLNSQRVNAVLYICFGSINYLSDKQLYEMACAIE 986
Query: 289 ASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEKGMILRG 330
S F+WVVP EK+GKE+E EEEKEKWLP+GFEE+ + G
Sbjct: 987 TSGHPFIWVVP-EKKGKEDESEEEKEKWLPKGFEERNISKMG 1109
>TC87789 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:flavonoid
glycosyltransferase {Vigna mungo}, partial (38%)
Length = 1081
Score = 328 bits (842), Expect = 2e-90
Identities = 166/265 (62%), Positives = 202/265 (75%), Gaps = 5/265 (1%)
Frame = +2
Query: 210 KHYERTTGGHKAWLLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCF 269
+HYE+ TG HK W LGP SL+R TA+EK+ERG+E V+ E LSWL +++ +SVLY+CF
Sbjct: 2 QHYEKATG-HKVWHLGPTSLIRKTAQEKSERGNE-GAVNVHESLSWLDSERVNSVLYICF 175
Query: 270 GSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEE-----K 324
GS+ FS++QL+EMAC IEAS F+WVVP EK+GKE+E EEEKEKWLP+GFEE K
Sbjct: 176 GSINYFSDKQLYEMACAIEASGHPFIWVVP-EKKGKEDESEEEKEKWLPKGFEERNIGKK 352
Query: 325 GMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEV 384
G+I+RGWAPQ+ IL HP+VG F+THCG NS +EAVSAGVP+ITWPV GDQFY EKL+T+
Sbjct: 353 GLIIRGWAPQVKILSHPAVGGFMTHCGGNSTVEAVSAGVPMITWPVHGDQFYNEKLITQF 532
Query: 385 RGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLAN 444
RG+GVEVGA EW G ++ ++SRD IEKAVRRLM GG+EAE IR R E G A
Sbjct: 533 RGIGVEVGATEWCTSGVAERKK--LVSRDSIEKAVRRLMDGGDEAENIRLRAREFGEKAI 706
Query: 445 AAVQPGGSSYQNLTALIAHLKTLRD 469
A+Q GGSSY NL ALI LK RD
Sbjct: 707 QAIQEGGSSYNNLLALIDELKRSRD 781
>TC86825 similar to GP|18151384|dbj|BAB83692. ABA-glucosyltransferase {Vigna
angularis}, partial (87%)
Length = 1704
Score = 305 bits (782), Expect = 2e-83
Identities = 181/479 (37%), Positives = 262/479 (53%), Gaps = 19/479 (3%)
Frame = +3
Query: 2 DVESEQRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPNLHL 61
++ E +K+YF P++ GH IP+ D A +FA G TIITTPSN +S+
Sbjct: 69 NMNHETDAVKIYFFPFVGGGHQIPMVDTARVFAKHGATSTIITTPSNALQFQNSITRDQK 248
Query: 62 HTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYCF 121
P + + P+ E + D+ + I L EP++ F+ H D +V D
Sbjct: 249 SNHPITIQILTTPENTEV--TDTDMSAGPMIDTSILL--EPLKQFLVQHRPDGIVVDMFH 416
Query: 122 PWIDDLTTELHIPRLSFNPSPLFALCAMRAKR----------GSSSFLISGLPHGDIALN 171
W D+ EL IPR+ FN + F C + R S F++ GLP
Sbjct: 417 RWAGDIIDELKIPRIVFNGNGCFPRCVIENTRKHVVLENLSSDSEPFIVPGLPDIIEMTR 596
Query: 172 ASPPEAL---TACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPAS 228
+ P + + + + + E S G LINSF +L+ Y R G KAWL+GP S
Sbjct: 597 SQTPIFMRNPSQFSDRIKQLEENSLGTLINSFYDLE--PAYADYVRNKLGKKAWLVGPVS 770
Query: 229 LVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIE 288
L + ++K ERG + ++ L+WL++KKP+SVLY+ FGS+ +QL E+A G+E
Sbjct: 771 LCNRSVEDKKERGKQPTI-DEQSCLNWLNSKKPNSVLYISFGSVARVPMKQLKEIAYGLE 947
Query: 289 ASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFE------EKGMILRGWAPQLLILGHPS 342
AS F+WVV GK + +E W+ + FE +KG+I RGWAPQLLIL H +
Sbjct: 948 ASDQSFIWVV-----GKILNSSKNEEDWVLDKFERRMKEMDKGLIFRGWAPQLLILEHEA 1112
Query: 343 VGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGR 402
VG F+THCG NS LE V AGVP+ TWP+ +QF EKL+T+V +GV+VG+ EW G
Sbjct: 1113VGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGSREW---GSW 1283
Query: 403 RERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALI 461
E + ++ R+K+E AV++LM E+ E++RRRV + A AV+ GGSSY ++ ALI
Sbjct: 1284DEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGGSSYDDIHALI 1460
>TC92399 weakly similar to GP|5763524|dbj|BAA83484.1 UDP-glucose: flavonoid
7-O-glucosyltransferase {Scutellaria baicalensis},
partial (29%)
Length = 843
Score = 291 bits (744), Expect = 5e-79
Identities = 146/239 (61%), Positives = 187/239 (78%), Gaps = 7/239 (2%)
Frame = +1
Query: 239 ERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVV 298
ERG E+SVVS E LSWL++K+ +SV+Y+CFGS C FS++QL+E+ACGIEASS EF+WVV
Sbjct: 1 ERG-EESVVSVHECLSWLNSKQDNSVVYICFGSQCHFSDKQLYEIACGIEASSHEFIWVV 177
Query: 299 PAEKRGKEEEEEEEKEKWLPEGFEE------KGMILRGWAPQLLILGHPSVGAFLTHCGS 352
P +KR E + EEEKEKWLP+GFEE K MI++GWAPQ++IL H +VGAF+THCG
Sbjct: 178 PEKKR-TENDNEEEKEKWLPKGFEERIIGKKKAMIIKGWAPQVMILSHTAVGAFMTHCGW 354
Query: 353 NSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGG-GRRERERVVIS 411
NS +EAVSAGVP+ITWP+ G+QFY EKL+T+V G+GVEVGA EWS G G RE+ V+
Sbjct: 355 NSTVEAVSAGVPMITWPMHGEQFYNEKLITQVHGIGVEVGATEWSTTGIGEREK---VVW 525
Query: 412 RDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHLKTLRDS 470
RD IEK V+RLM G+EAE+IR+ E G A A++ GGSS+ NLTA++ +LK LRD+
Sbjct: 526 RDNIEKVVKRLMDSGDEAEKIRQHAREFGEKAKHAIKEGGSSHSNLTAVVNYLKRLRDN 702
>TC80062 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:flavonoid
glycosyltransferase {Vigna mungo}, partial (42%)
Length = 1071
Score = 272 bits (695), Expect = 2e-73
Identities = 140/229 (61%), Positives = 176/229 (76%), Gaps = 6/229 (2%)
Frame = +2
Query: 247 VSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKE 306
VS E LSWL +K+ +SVLY+CFGS+ FS++QL+E+A GIE S +FVWVVP EK+GKE
Sbjct: 2 VSVHECLSWLDSKEDNSVLYICFGSISHFSDKQLYEIASGIENSGHKFVWVVP-EKKGKE 178
Query: 307 EEEEEEKEKWLPEGFEE------KGMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVS 360
+E EE+KEKWLP+GFEE KG I++GWAPQ +IL H VGAF+THCG NS +EA+S
Sbjct: 179 DESEEQKEKWLPKGFEERNILNKKGFIIKGWAPQAMILSHTVVGAFMTHCGWNSIVEAIS 358
Query: 361 AGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVR 420
AG+P+ITWPV G+QFY EKL+T V+ +GVEVGA EWS+ G +E+E+VV SR IEKAVR
Sbjct: 359 AGIPMITWPVHGEQFYNEKLITVVQRIGVEVGATEWSL-HGFQEKEKVV-SRHSIEKAVR 532
Query: 421 RLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHLKTLRD 469
RLM G+EA++IR+R E G A AV+ GGSS+ NL ALI LK RD
Sbjct: 533 RLMDDGDEAKEIRQRAQEFGKKATHAVEEGGSSHNNLLALINDLKRSRD 679
>BG450101 similar to GP|4115534|dbj UDP-glycose:flavonoid glycosyltransferase
{Vigna mungo}, partial (21%)
Length = 646
Score = 234 bits (598), Expect(2) = 6e-64
Identities = 116/198 (58%), Positives = 147/198 (73%), Gaps = 5/198 (2%)
Frame = +1
Query: 187 RKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVRGTAKEKAERGHEKSV 246
RK K L + DG EY+K+Y +TG HKAW LGPASL+R T +EKAERG E S
Sbjct: 34 RKSSKVMDSLSTISLSXDGEEYIKYYVSSTG-HKAWHLGPASLIRKTVQEKAERGQE-SA 207
Query: 247 VSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKE 306
VS E LSWL++K+ +SVLY+ FGS+C + ++QL+E+AC IEAS +F+WV+P GKE
Sbjct: 208 VSVQECLSWLNSKRHNSVLYISFGSLCRYQDKQLYEIACAIEASGHDFIWVIPLNN-GKE 384
Query: 307 EEEEEEKEKWLPEGFEE-----KGMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSA 361
+E EEEK+KWLP+GFEE KG+I+RGWAPQ+LIL HP+VGAF+THCG NS +EAV A
Sbjct: 385 DESEEEKQKWLPKGFEERNIGKKGLIIRGWAPQVLILSHPAVGAFMTHCGWNSTVEAVGA 564
Query: 362 GVPIITWPVLGDQFYTEK 379
GVP+ITWP G+QF K
Sbjct: 565 GVPMITWPSHGEQFLHRK 618
Score = 28.1 bits (61), Expect(2) = 6e-64
Identities = 11/16 (68%), Positives = 14/16 (86%)
Frame = +2
Query: 373 DQFYTEKLMTEVRGVG 388
+ FYTEKL+TEVR +G
Sbjct: 599 NNFYTEKLITEVRXIG 646
>BQ165264 weakly similar to PIR|T07786|T077 UDP-glucose glucosyltransferase
(EC 2.4.1.-) - potato, partial (7%)
Length = 847
Score = 238 bits (608), Expect = 3e-63
Identities = 132/262 (50%), Positives = 168/262 (63%), Gaps = 15/262 (5%)
Frame = +2
Query: 7 QRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPN-----LHL 61
+RPLKLY +P+L+ GHMIPL DIA LFAS GQ VTIITTPSN SL + L L
Sbjct: 68 ERPLKLYLLPFLSPGHMIPLGDIATLFASHGQQVTIITTPSNAHFFTKSLSSVDPFFLRL 247
Query: 62 HTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYCF 121
HTV FPS+QVGLPDG+ES SS+ D TT K++ G LL PI+ F+E P D ++ D F
Sbjct: 248 HTVDFPSQQVGLPDGVESLSSNIDTDTTHKIYVGSMLLHGPIKEFIEKDPPDYIIGDCVF 427
Query: 122 PWIDDLTTELHIPRLSFNPSPLFALCAMRAKR----------GSSSFLISGLPHGDIALN 171
PWI DL + HI L+F LF++ + A R SSSF++ PH I N
Sbjct: 428 PWIHDLANKPHISTLAFTGYSLFSVSLIEALRVHRSNSHTNSDSSSFVVPNFPH-SITFN 604
Query: 172 ASPPEALTACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVR 231
+ PP+ E +L+ +KS G++IN+FVELDG + +KHYE+T GHKAW LGPA L+
Sbjct: 605 SGPPKTFIEFEEGMLKTIIKSKGLIINNFVELDGEDCIKHYEKTM-GHKAWHLGPACLIH 781
Query: 232 GTAKEKAERGHEKSVVSTDELL 253
+ +EKAERG+E SVVS E L
Sbjct: 782 ESVQEKAERGNE-SVVSMHECL 844
>TC79031 weakly similar to GP|13492674|gb|AAK28303.1
phenylpropanoid:glucosyltransferase 1 {Nicotiana
tabacum}, partial (32%)
Length = 1385
Score = 230 bits (587), Expect = 8e-61
Identities = 147/402 (36%), Positives = 217/402 (53%), Gaps = 24/402 (5%)
Frame = +1
Query: 91 KVHQGIKLLREPIQLFMEHHPADCVVADYCFPWIDDLTTELHIPRLSFNPSPLFALCA-- 148
K++ G+ +L+ I+ E D +V D FPW D+ +L IPR+ F+ + A A
Sbjct: 28 KIYMGLYILQPDIEKLFETLQPDFIVTDMFFPWSADVAKKLGIPRIMFHGASYLARSAAH 207
Query: 149 --------MRAKRGSSSFLISGLPHGDIALNASPPEALTA------CVEPLLRKELKSHG 194
++A+ + F+I LP P+ L + ++ + E KS G
Sbjct: 208 SVEVYRPHLKAESDTDKFVIPDLPDELEMTRLQLPDWLRSPNQYAELMKVIKESEKKSFG 387
Query: 195 VLINSFVELDGHEYVKHYERTTGGHKAWLLGPASL-VRGTAKEKAERGHEKSVVSTDE-- 251
+ NSF +L+ EY HY++ G K+W LGP SL +KA RG+ + E
Sbjct: 388 SVFNSFYKLES-EYYDHYKKVMGT-KSWGLGPVSLWANQDDSDKAARGYARKEEGAKEEG 561
Query: 252 LLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEE- 310
L WL++K SVLYV FGSM F QL E+A +E S F+WVV R EE EE
Sbjct: 562 WLKWLNSKPDGSVLYVSFGSMNKFPYSQLVEIAHALENSGHNFIWVV----RKNEENEEG 729
Query: 311 ----EEKEKWLPEGFEEKGMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPII 366
EE EK + E KG ++ GWAPQLLIL + ++G ++HCG N+ +E+V+ G+P +
Sbjct: 730 GVFLEEFEKKMKES--GKGYLIWGWAPQLLILENHAIGGLVSHCGWNTVVESVNVGLPTV 903
Query: 367 TWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGG 426
TWP+ + F+ EKL+ +V +GV VGA+EW E V+ R+ I A+R +M GG
Sbjct: 904 TWPLFAEHFFNEKLVVDVLKIGVPVGAKEWR---NWNEFGSEVVKREDIGNAIRLMMEGG 1074
Query: 427 EEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHLKTLR 468
EE +R+RV EL A A++ GGSSY N+ LI L++++
Sbjct: 1075EEEVAMRKRVKELSVEAKKAIKVGGSSYNNMVELIQELRSIK 1200
>TC79293 weakly similar to GP|15290056|dbj|BAB63750. putative
glucosyltransferase IS5a salicylate-induced {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 866
Score = 222 bits (566), Expect = 2e-58
Identities = 126/269 (46%), Positives = 169/269 (61%), Gaps = 15/269 (5%)
Frame = +3
Query: 7 QRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPN-----LHL 61
+RPLKL+ +P+L+ GHMIPL DIA LFAS GQ VTIITTPSN D S+ + L L
Sbjct: 69 ERPLKLHMLPFLSPGHMIPLGDIAALFASHGQQVTIITTPSNAHFFDKSIASVDPFFLRL 248
Query: 62 HTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYCF 121
H V FPS+QV LPDG+ES SS+ T K+ +G LL EPI+ F+E D ++AD +
Sbjct: 249 HIVDFPSQQVDLPDGVESLSSTTGPATMAKICKGANLLHEPIREFVEKDQPDYIIADCVY 428
Query: 122 PWIDDLTTELHIPRLSFNPSPLFALCAMRAKR----------GSSSFLISGLPHGDIALN 171
PWI+DLT + HI ++F LF + + + R SSF+ PH I
Sbjct: 429 PWINDLTNKPHISTIAFTGFSLFTISLIESLRINRSYFDKNSSLSSFVDPNFPH-SITFC 605
Query: 172 ASPPEALTACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVR 231
A+ P+ L A E +L KS G+++N+F ELDG + +KHYE+T G+KAW LGPA L+R
Sbjct: 606 ATTPKQLIAFEERMLETIRKSKGLIVNNFAELDGEDCIKHYEKTM-GYKAWHLGPACLIR 782
Query: 232 GTAKEKAERGHEKSVVSTDELLSWLSTKK 260
T +EK+ RG+E SVVS E SWL++K+
Sbjct: 783 KTFQEKSVRGNE-SVVSVHECPSWLNSKE 866
>AW559311 weakly similar to PIR|T03747|T037 glucosyltransferase IS5a (EC
2.4.1.-) salicylate-induced - common tobacco, partial
(18%)
Length = 572
Score = 213 bits (542), Expect = 1e-55
Identities = 109/193 (56%), Positives = 144/193 (74%), Gaps = 6/193 (3%)
Frame = +2
Query: 174 PPEALTACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVRGT 233
PP+ A + +L KS G++ N+FVEL + +KHYE+T G +KAW LGP L+R T
Sbjct: 2 PPKLYVAFEDRILEAIRKSKGLIFNNFVELPREDCIKHYEKTMG-YKAWHLGPTCLIRKT 178
Query: 234 AKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVE 293
+EK+ RG+E SVVS + L WL++K+ +SVLY+CFGS+ FS++QL+E+A GIE S E
Sbjct: 179 FEEKSMRGNE-SVVSAHKCLGWLNSKQDNSVLYICFGSISYFSDKQLYEIASGIENSGHE 355
Query: 294 FVWVVPAEKRGKEEEEEEEKEKWLPEGFEE------KGMILRGWAPQLLILGHPSVGAFL 347
FVWVVP EK+GK++E EEEKEKWLP+GFEE KG I+R WAPQ++IL H +VGAF+
Sbjct: 356 FVWVVP-EKKGKKDESEEEKEKWLPKGFEERNVENKKGFIIREWAPQVMILSHAAVGAFM 532
Query: 348 THCGSNSALEAVS 360
THCGSNS +EAVS
Sbjct: 533 THCGSNSLVEAVS 571
>BG645363 weakly similar to GP|15290080|dbj putative salicylate-induced
glucosyltransferase {Oryza sativa (japonica
cultivar-group)}, partial (9%)
Length = 796
Score = 209 bits (531), Expect = 2e-54
Identities = 121/250 (48%), Positives = 156/250 (62%), Gaps = 15/250 (6%)
Frame = +3
Query: 7 QRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPN-----LHL 61
+RPLKL +P+LA GHMIPL DIA LFAS GQ VTIITTPSN SL L L
Sbjct: 30 ERPLKLLILPFLAPGHMIPLGDIAALFASHGQQVTIITTPSNAHFFTKSLSFVDPFFLRL 209
Query: 62 HTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYCF 121
HTV FPS+QV LPDG+ES SS+ + T K+ +G LL EPI+ F+E D ++AD +
Sbjct: 210 HTVDFPSQQVDLPDGVESLSSTTNPDTMAKICKGAMLLHEPIREFVEKDQPDYIIADCVY 389
Query: 122 PWIDDLTTELHIPRLSFNPSPLFALC---AMRAKR-------GSSSFLISGLPHGDIALN 171
PWI+DL + HI ++F P LF + ++R KR SSSF+ PH I
Sbjct: 390 PWINDLANKPHISTIAFTPFFLFTVSLIESLRIKRSYSDKNSSSSSFVDPNFPH-SINFC 566
Query: 172 ASPPEALTACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVR 231
++PP+ A E +L KS G++IN+FVELDG + +KHYE+T G+KAW LGP L+R
Sbjct: 567 STPPKLYVAFEERMLEAIRKSKGLIINNFVELDGEDCIKHYEKTM-GYKAWHLGPTCLIR 743
Query: 232 GTAKEKAERG 241
T EK+ RG
Sbjct: 744 KTF*EKSMRG 773
>BQ123034 weakly similar to GP|14334982|gb putative glucosyltransferase
{Arabidopsis thaliana}, partial (24%)
Length = 765
Score = 186 bits (473), Expect = 1e-47
Identities = 102/236 (43%), Positives = 148/236 (62%), Gaps = 6/236 (2%)
Frame = +2
Query: 244 KSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKR 303
KS+ ELL+WL++K+ SVLYV FGS QL E+ G+E S F+WV+
Sbjct: 53 KSLGKHTELLNWLNSKENESVLYVSFGSFTRLPYAQLVEIVHGLENSGHNFIWVI----- 217
Query: 304 GKEEEEEEEKEKWLPEGFEE------KGMILRGWAPQLLILGHPSVGAFLTHCGSNSALE 357
K ++ +E+ E +L E FEE KG I+ WAPQLLIL HP+ G +THCG NS LE
Sbjct: 218 -KRDDTDEDGEGFLQE-FEERIKESSKGYIIWDWAPQLLILDHPATGGIVTHCGWNSTLE 391
Query: 358 AVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEK 417
+++AG+P+ITWP+ +QFY EKL+ +V +GV VGA+E + + V+ R++IEK
Sbjct: 392 SLNAGLPMITWPIFAEQFYNEKLLVDVLKIGVPVGAKENKLWLDISVEK--VVRREEIEK 565
Query: 418 AVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHLKTLRDSNSV 473
V+ LMG G+E++++R R +L A ++ GG SY NL LI LK+L+ S ++
Sbjct: 566 TVKILMGSGQESKEMRMRAKKLSEAAKRTIEEGGDSYNNLIQLIDELKSLKKSKAL 733
>TC78545 weakly similar to GP|13492676|gb|AAK28304.1
phenylpropanoid:glucosyltransferase 2 {Nicotiana
tabacum}, partial (10%)
Length = 589
Score = 178 bits (452), Expect = 4e-45
Identities = 97/210 (46%), Positives = 128/210 (60%), Gaps = 2/210 (0%)
Frame = +2
Query: 7 QRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPN--LHLHTV 64
+RPLKL+FIPY A+GHM+PLCDIA LFASRGQHVTIITTPSN ++ +L + L LHTV
Sbjct: 62 ERPLKLHFIPYPASGHMMPLCDIATLFASRGQHVTIITTPSNAQSLTKTLSSAALRLHTV 241
Query: 65 PFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYCFPWI 124
FP +QV LP G+ES +S+ D TT+K+H G LL E + F+E +P DC++AD
Sbjct: 242 EFPYQQVDLPKGVESMTSTTDPITTWKIHNGAMLLNEAVGDFVEKNPPDCIIAD------ 403
Query: 125 DDLTTELHIPRLSFNPSPLFALCAMRAKRGSSSFLISGLPHGDIALNASPPEALTACVEP 184
SSS+++ L H +I L + PP+ L+ +
Sbjct: 404 ----------------------------SDSSSYVVPNLHHDNITLCSKPPKVLSMFIGM 499
Query: 185 LLRKELKSHGVLINSFVELDGHEYVKHYER 214
+L LKS G +IN+FVELDG E VKHYE+
Sbjct: 500 MLDTVLKSTGYIINNFVELDGEECVKHYEK 589
>TC78653 similar to PIR|T46162|T46162 glucosyltransferase-like protein -
Arabidopsis thaliana, partial (6%)
Length = 712
Score = 177 bits (449), Expect = 8e-45
Identities = 94/194 (48%), Positives = 127/194 (65%), Gaps = 18/194 (9%)
Frame = +1
Query: 3 VESEQRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPN---- 58
VE EQ PLK+YFIP+LA+GHMIPL DIA +FASRGQ VT+ITTP+N ++ SL +
Sbjct: 112 VEVEQ-PLKVYFIPFLASGHMIPLFDIATMFASRGQQVTVITTPANAKSLTKSLSSDAPS 288
Query: 59 -LHLHTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVA 117
L LHTV FPS+QVGLP+GIES SS+ D TT+K+H G LL+EPI F+E+ P DC+++
Sbjct: 289 FLRLHTVDFPSQQVGLPEGIESMSSTTDPTTTWKIHTGAMLLKEPIGDFIENDPPDCIIS 468
Query: 118 DYCFPWIDDLTTELHIPRLSFNPSPLFALCAMRAKR-------------GSSSFLISGLP 164
D +PW++DL + IP ++FN LFA+ + + SSSF++ P
Sbjct: 469 DSTYPWVNDLADKFQIPNITFNGLCLFAVSLVETLKTNNLLKSQTDSDSDSSSFVVPNFP 648
Query: 165 HGDIALNASPPEAL 178
H I L PP+ +
Sbjct: 649 H-HITLCGKPPKVI 687
>AJ502218 similar to GP|22208467|gb| putative glucosyl transferase {Sorghum
bicolor}, partial (5%)
Length = 680
Score = 177 bits (448), Expect = 1e-44
Identities = 98/204 (48%), Positives = 123/204 (60%), Gaps = 16/204 (7%)
Frame = +1
Query: 8 RPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPN-----LHLH 62
RP+KL+FIPY A GHMIP+CDIA LFAS GQ VTIITTPSN+ SL + L LH
Sbjct: 70 RPMKLHFIPYPAPGHMIPMCDIATLFASHGQQVTIITTPSNSHFFTKSLSSVDPFFLRLH 249
Query: 63 TVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYCFP 122
V FPS+QVGLPDG+ES SS++D T+ K++ G LLREPI FME P DC++AD P
Sbjct: 250 LVNFPSQQVGLPDGVESMSSTSDGDTSGKIYYGAMLLREPIHDFMEKDPPDCIIADCINP 429
Query: 123 WIDDLTTELHIPRLSFNPSPLFALCAMRAKR-----------GSSSFLISGLPHGDIALN 171
W+ DL +L IP ++F LF + M + R SF + PH I L
Sbjct: 430 WVYDLAHKLQIPTIAFTGFSLFTVSLMESLRINPSFCSHTDSSPGSFTVPNFPH-SITLC 606
Query: 172 ASPPEALTACVEPLLRKELKSHGV 195
PP+ T +E +L LKS G+
Sbjct: 607 TKPPKIFTGFMEMMLETILKSKGL 678
>TC78003 similar to GP|20146093|dbj|BAB88935. glucosyltransferase NTGT2
{Nicotiana tabacum}, partial (50%)
Length = 1740
Score = 168 bits (425), Expect = 5e-42
Identities = 149/482 (30%), Positives = 227/482 (46%), Gaps = 29/482 (6%)
Frame = +3
Query: 9 PLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPS--NTTTIDSSLPNLHLHTVPF 66
P ++ +PY GH+ P + A + G HVTI TT N T +LPNL + PF
Sbjct: 144 PHRILLVPYPVQGHINPAFEFAKRLITLGAHVTISTTVHMHNRITNKPTLPNLSYY--PF 317
Query: 67 PSRQVGLPDGIESFSSSADLQTTFKVHQ-GIKLLREPI-QLFMEHHPADCVVADYCFPWI 124
G DG + S A L+ + + G + + + I + E P C+V W
Sbjct: 318 SD---GYDDGFKGTGSDAYLEYHAEFQRRGSEFVSDIILKNSQEGTPFTCLVHSLLLQWA 488
Query: 125 DDLTTELHIPR--LSFNPSPLFALC-------AMRAKRGSSSFLISGLP----HGDIA-- 169
+ E H+P L P+ +F + + K SSS + GLP D+
Sbjct: 489 AEAAREFHLPTALLWVQPATVFDILYYYFHGFSDSIKNPSSSIELPGLPLLFSSRDLPSF 668
Query: 170 LNASPPEALTACV----EPLLRKELKSH---GVLINSFVELDGHEY--VKHYERTTGGHK 220
L AS P+A + E +++++ +L+NSF L+ VK + + G
Sbjct: 669 LLASCPDAYSLMTSFFEEQFNELDVETNLTKTILVNSFESLEPKALRAVKKFNMISIGP- 845
Query: 221 AWLLGPASLVRGTAKEKAERGHEKSVVS-TDELLSWLSTKKPSSVLYVCFGSMCSFSNEQ 279
L+ L + E G + + +++ + WL +K SSV+YV FGS S Q
Sbjct: 846 --LIPSEHLDEKDSTEDNSYGGQTHIFQPSNDCVEWLDSKPKSSVVYVSFGSYFVLSERQ 1019
Query: 280 LHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEKGMILRGWAPQLLILG 339
E+A + F+WV+ R KE E EE K+ E EEKG I++ W Q+ IL
Sbjct: 1020REEIAHALLDCGFPFLWVL----REKEGENNEEGFKYREE-LEEKGKIVK-WCSQMEILS 1181
Query: 340 HPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMG 399
HPS+G FLTHCG NS LE++ GVP++ +P DQ KL+ +V +GV V E
Sbjct: 1182HPSLGCFLTHCGWNSTLESLVKGVPMVAFPQWTDQMTNAKLIEDVWKIGVRVDEE----- 1346
Query: 400 GGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTA 459
E ++ D+I + + +MG GE+ E++RR + LA AV+ GGSS +NL +
Sbjct: 1347----VNEDGIVRGDEIRRCLEVVMGSGEKGEELRRSGKKWKELAREAVKEGGSSEKNLRS 1514
Query: 460 LI 461
+
Sbjct: 1515FL 1520
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,020,328
Number of Sequences: 36976
Number of extensions: 246235
Number of successful extensions: 2336
Number of sequences better than 10.0: 193
Number of HSP's better than 10.0 without gapping: 1818
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1985
length of query: 481
length of database: 9,014,727
effective HSP length: 100
effective length of query: 381
effective length of database: 5,317,127
effective search space: 2025825387
effective search space used: 2025825387
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0367.1


