

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
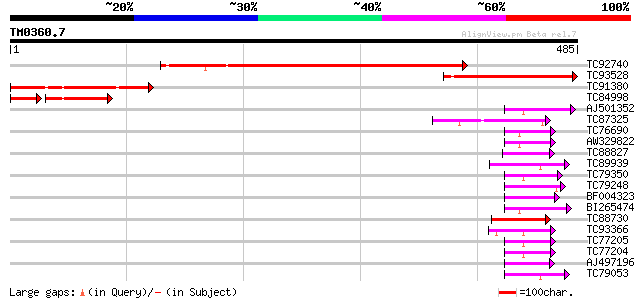
Query= TM0360.7
(485 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC92740 similar to GP|20466490|gb|AAM20562.1 unknown protein {Ar... 412 e-115
TC93528 similar to GP|20466490|gb|AAM20562.1 unknown protein {Ar... 194 6e-50
TC91380 similar to GP|10726444|gb|AAG22141.1 CG4655-PA {Drosophi... 136 2e-32
TC84998 similar to GP|19915277|gb|AAM04835.1 cell surface protei... 81 5e-19
AJ501352 similar to PIR|B84584|B845 probable RING zinc finger pr... 54 1e-07
TC87325 similar to GP|18176187|gb|AAL60000.1 unknown protein {Ar... 53 3e-07
TC76690 similar to GP|21593806|gb|AAM65773.1 putative RING zinc ... 51 9e-07
AW329822 similar to PIR|T00428|T004 hypothetical protein At2g475... 51 9e-07
TC88827 similar to GP|22165059|gb|AAM93676.1 unknown protein {Or... 48 7e-06
TC89939 similar to GP|15289911|dbj|BAB63606. hypothetical protei... 48 7e-06
TC79350 similar to PIR|T08944|T08944 hypothetical protein F25O24... 48 1e-05
TC79248 similar to GP|22165059|gb|AAM93676.1 unknown protein {Or... 48 1e-05
BF004323 similar to GP|4337011|gb| zinc-binding peroxisomal inte... 47 1e-05
BI265474 weakly similar to PIR|T48058|T480 RING-H2 zinc finger p... 47 2e-05
TC88730 weakly similar to GP|21592748|gb|AAM64697.1 PRT1 {Arabid... 46 3e-05
TC93366 similar to PIR|E96745|E96745 hypothetical protein T9N14.... 46 4e-05
TC77205 similar to GP|19347753|gb|AAL86301.1 putative RING-H2 zi... 46 4e-05
TC77204 similar to GP|21536625|gb|AAM60957.1 RING-H2 zinc finger... 46 4e-05
AJ497196 weakly similar to SP|O60683|PEXA_ Peroxisome assembly p... 45 6e-05
TC79053 similar to GP|22795037|gb|AAN05420.1 putative RING prote... 45 8e-05
>TC92740 similar to GP|20466490|gb|AAM20562.1 unknown protein {Arabidopsis
thaliana}, partial (46%)
Length = 789
Score = 412 bits (1060), Expect = e-115
Identities = 210/265 (79%), Positives = 231/265 (86%), Gaps = 3/265 (1%)
Frame = +3
Query: 130 LMGERRGAGMSPL-DDDPEGRGGAAVNETVPLVSSSAS--LAGSVQGDGDAAGNGAESDS 186
L+ ER +GM L ++D EGRGG +E VPLVSSS+S LAGS Q DG+AAGNG E++
Sbjct: 3 LIAER--SGMPGLGENDAEGRGGIEASEGVPLVSSSSSSSLAGSGQVDGEAAGNGTENN- 173
Query: 187 RGSSSYQRYDIQQVAKWIEQILPFSLLLLIVFIRQHLQGFFVTIWISAVMFKSNEIVKKQ 246
R SSSYQRYDIQ VAKWIEQILPFSLLLL+VFIRQHLQGFFVTIWI+AVMFKSNEIVKKQ
Sbjct: 174 RDSSSYQRYDIQLVAKWIEQILPFSLLLLVVFIRQHLQGFFVTIWIAAVMFKSNEIVKKQ 353
Query: 247 TALKGDRKVSVLVGISFAFILQVTCIYWWYRNDDLLYPLIMLPPRPTPFWHSVFIILVND 306
TALKGDRKVS+L GI+FAFIL V CIYWWYRNDD+LYPL MLPP PTPFWH++F ILVND
Sbjct: 354 TALKGDRKVSLLAGIAFAFILHVICIYWWYRNDDILYPLAMLPPNPTPFWHAIFTILVND 533
Query: 307 VLVRQAAMVFKCFLLIYYKNGKGHNFRRQAQMLTLVEYMLLLYRALLPTLVWYRFFLNRD 366
+L+RQ AM FKC LLIYYK GKGHNFRRQAQMLTLVEY LLLYRALLPT VWYRFFLNR+
Sbjct: 534 ILMRQVAMAFKCILLIYYKKGKGHNFRRQAQMLTLVEYTLLLYRALLPTPVWYRFFLNRE 713
Query: 367 YGSLFSSLTTGLYLTFKLTSIIEKV 391
YGSLFSSLTTGLYLTFKL ++ KV
Sbjct: 714 YGSLFSSLTTGLYLTFKLHLVVXKV 788
>TC93528 similar to GP|20466490|gb|AAM20562.1 unknown protein {Arabidopsis
thaliana}, partial (24%)
Length = 755
Score = 194 bits (493), Expect = 6e-50
Identities = 94/114 (82%), Positives = 102/114 (89%)
Frame = +3
Query: 372 SSLTTGLYLTFKLTSIIEKVRGFISAFKALSRKDVHYGVYATMEQVNAAGDLCAICQEKM 431
SSLTTG + F+ +EKV+ FISA KALSRK+VHYGVYAT EQV AAGDLCAICQEKM
Sbjct: 15 SSLTTGC-IYFQANICVEKVQCFISALKALSRKEVHYGVYATAEQVTAAGDLCAICQEKM 191
Query: 432 HSPVLLRCKHIFCEECVSEWFERERTCPLCRALVKPADLQTFGDGSTSLFFQLF 485
HSP+LLRCKHIFCE+CVSEWFERERTCPLCRALVK ADL+TFGDGSTSLFFQLF
Sbjct: 192 HSPILLRCKHIFCEDCVSEWFERERTCPLCRALVKAADLRTFGDGSTSLFFQLF 353
>TC91380 similar to GP|10726444|gb|AAG22141.1 CG4655-PA {Drosophila
melanogaster}, partial (3%)
Length = 687
Score = 136 bits (342), Expect = 2e-32
Identities = 78/123 (63%), Positives = 87/123 (70%)
Frame = +3
Query: 1 METLDGNSDQYATVSSNSPSSSSGGFNSSDDHNNGSVSWYGMRLPSVNPLRSPLSLLLDY 60
METL GNS ++SS S SSS GG N+ D NNGSVSWYGMRLP VNP SP+S LLDY
Sbjct: 273 METLGGNSSPSPSLSSASSSSSFGGINTDD--NNGSVSWYGMRLP-VNPFLSPISFLLDY 443
Query: 61 SGILTQTHDSGSVIVNNGVPASEIRSQVQMQPFDDAVGGSAGEVAIRIIGAGEHDQNQIG 120
SGIL +DS +IVNNGV SE+R QV G SAGEVAIRIIGAGE+ NQ+G
Sbjct: 444 SGILRSGNDSEGMIVNNGVSGSELRPQVDAG--GAVAGSSAGEVAIRIIGAGENIHNQVG 617
Query: 121 EVG 123
EVG
Sbjct: 618 EVG 626
>TC84998 similar to GP|19915277|gb|AAM04835.1 cell surface protein
{Methanosarcina acetivorans str. C2A} [Methanosarcina
acetivorans C2A], partial (3%)
Length = 622
Score = 81.3 bits (199), Expect(2) = 5e-19
Identities = 41/58 (70%), Positives = 45/58 (76%)
Frame = +1
Query: 31 DHNNGSVSWYGMRLPSVNPLRSPLSLLLDYSGILTQTHDSGSVIVNNGVPASEIRSQV 88
D NNGSVSWYGMRLP VNP SP+S LLDYSGIL +DS +IVNNGV SE+R QV
Sbjct: 442 DDNNGSVSWYGMRLP-VNPFLSPISFLLDYSGILRSGNDSEGMIVNNGVSGSELRPQV 612
Score = 31.2 bits (69), Expect(2) = 5e-19
Identities = 16/27 (59%), Positives = 18/27 (66%)
Frame = +3
Query: 1 METLDGNSDQYATVSSNSPSSSSGGFN 27
METL GNS ++SS S SSS GG N
Sbjct: 357 METLGGNSSPSPSLSSASSSSSFGGIN 437
>AJ501352 similar to PIR|B84584|B845 probable RING zinc finger protein
[imported] - Arabidopsis thaliana, partial (17%)
Length = 607
Score = 54.3 bits (129), Expect = 1e-07
Identities = 23/65 (35%), Positives = 35/65 (53%), Gaps = 4/65 (6%)
Frame = +3
Query: 424 CAICQEKMHSPVLLR----CKHIFCEECVSEWFERERTCPLCRALVKPADLQTFGDGSTS 479
C +C K +LR CKH F C+ +W E TCPLCR +V+ +D++ F +S
Sbjct: 411 CTVCLSKFEDEEILRLLPKCKHAFHMNCIDKWLESHSTCPLCRYMVEESDIRNFTFSFSS 590
Query: 480 LFFQL 484
F ++
Sbjct: 591 RFLRV 605
>TC87325 similar to GP|18176187|gb|AAL60000.1 unknown protein {Arabidopsis
thaliana}, partial (50%)
Length = 1568
Score = 52.8 bits (125), Expect = 3e-07
Identities = 34/108 (31%), Positives = 55/108 (50%), Gaps = 7/108 (6%)
Frame = +3
Query: 362 FLNRDYGSLFSSLTTGLYLTFK--LTSIIEKVRGFISAFKALSRKDVHYGVY-ATMEQVN 418
++ R +G F + L+L + L+++I +++GFI AL +H + AT E++
Sbjct: 147 YIWRLHGMAFHLVDAVLFLNIRALLSAMINRIKGFIRLRIALGA--LHAALPDATTEELR 320
Query: 419 AAGDLCAICQEKMHSPVLLRCKHIFCEECVSEWFER----ERTCPLCR 462
D CAIC+E M L C H+F C+ W ++ TCP CR
Sbjct: 321 GYEDECAICREPMAKAKKLNCNHLFHLACLRSWLDQGLTEMYTCPTCR 464
>TC76690 similar to GP|21593806|gb|AAM65773.1 putative RING zinc finger
protein {Arabidopsis thaliana}, partial (22%)
Length = 1303
Score = 51.2 bits (121), Expect = 9e-07
Identities = 23/48 (47%), Positives = 28/48 (57%), Gaps = 4/48 (8%)
Frame = +3
Query: 424 CAICQEKMHSP----VLLRCKHIFCEECVSEWFERERTCPLCRALVKP 467
CA+C + S VL +CKH F EC+ WF TCPLCRA V+P
Sbjct: 381 CAVCLSEFESGETGRVLPKCKHSFHTECIDMWFHSHDTCPLCRAPVEP 524
>AW329822 similar to PIR|T00428|T004 hypothetical protein At2g47560
[imported] - Arabidopsis thaliana, partial (27%)
Length = 553
Score = 51.2 bits (121), Expect = 9e-07
Identities = 23/48 (47%), Positives = 28/48 (57%), Gaps = 4/48 (8%)
Frame = +1
Query: 424 CAICQEKMHSP----VLLRCKHIFCEECVSEWFERERTCPLCRALVKP 467
CA+C + S VL +CKH F EC+ WF TCPLCRA V+P
Sbjct: 247 CAVCLSEFESGETGRVLPKCKHSFHTECIDMWFHSHDTCPLCRAPVEP 390
>TC88827 similar to GP|22165059|gb|AAM93676.1 unknown protein {Oryza sativa
(japonica cultivar-group)}, complete
Length = 1221
Score = 48.1 bits (113), Expect = 7e-06
Identities = 19/45 (42%), Positives = 24/45 (53%)
Frame = +3
Query: 422 DLCAICQEKMHSPVLLRCKHIFCEECVSEWFERERTCPLCRALVK 466
D C IC E VL C H+ C +C EW R ++CP CR +K
Sbjct: 582 DECGICMEMNSKIVLPNCNHVMCLKCYREWRTRSQSCPFCRDSLK 716
>TC89939 similar to GP|15289911|dbj|BAB63606. hypothetical protein~similar
to Arabidopsis thaliana chromosome 1 F18O14.3, partial
(23%)
Length = 978
Score = 48.1 bits (113), Expect = 7e-06
Identities = 23/74 (31%), Positives = 40/74 (53%), Gaps = 5/74 (6%)
Frame = +2
Query: 411 YATMEQVNAAGDL-CAICQEKMHSPVLLRCKHIFCEECVSEWF---ERERTCPLCRALVK 466
Y++ + AG+ C IC + P++ C H+FC C+ +W + R CP+C+A+++
Sbjct: 104 YSSNNNNSDAGNFECNICFDLAQDPIITLCGHLFCWPCLYKWLHFHSQSRECPVCKAMIE 283
Query: 467 PADL-QTFGDGSTS 479
L +G G TS
Sbjct: 284 EEKLVPLYGRGKTS 325
>TC79350 similar to PIR|T08944|T08944 hypothetical protein F25O24.10 -
Arabidopsis thaliana, partial (32%)
Length = 1698
Score = 47.8 bits (112), Expect = 1e-05
Identities = 21/54 (38%), Positives = 28/54 (50%), Gaps = 4/54 (7%)
Frame = +3
Query: 424 CAICQEKMHSPVLLR----CKHIFCEECVSEWFERERTCPLCRALVKPADLQTF 473
C+IC K +LR CKH F +C+ W E+ +CP+CR V D TF
Sbjct: 474 CSICLSKFEDIEILRLLPKCKHAFHIDCIDHWLEKHSSCPICRHKVNIEDQTTF 635
>TC79248 similar to GP|22165059|gb|AAM93676.1 unknown protein {Oryza sativa
(japonica cultivar-group)}, partial (97%)
Length = 1272
Score = 47.8 bits (112), Expect = 1e-05
Identities = 22/55 (40%), Positives = 27/55 (49%), Gaps = 3/55 (5%)
Frame = +2
Query: 424 CAICQEKMHSPVLLRCKHIFCEECVSEWFERERTCPLCRALVK---PADLQTFGD 475
C IC E VL C H+ C +C EW R ++CP CR +K DL F D
Sbjct: 560 CGICMEMNSKIVLPNCNHVMCLKCYHEWRARSQSCPFCRDSLKRVNSGDLWIFTD 724
>BF004323 similar to GP|4337011|gb| zinc-binding peroxisomal integral
membrane protein {Arabidopsis thaliana}, partial (32%)
Length = 513
Score = 47.4 bits (111), Expect = 1e-05
Identities = 16/47 (34%), Positives = 23/47 (48%)
Frame = -1
Query: 424 CAICQEKMHSPVLLRCKHIFCEECVSEWFERERTCPLCRALVKPADL 470
C +C P C H+FC C++EW + CPLCR + + L
Sbjct: 255 CTLCLSNRQHPTATSCGHVFCWNCITEWCNEKPECPLCRTPITHSSL 115
>BI265474 weakly similar to PIR|T48058|T480 RING-H2 zinc finger protein ATL5
- Arabidopsis thaliana, partial (19%)
Length = 617
Score = 47.0 bits (110), Expect = 2e-05
Identities = 23/61 (37%), Positives = 30/61 (48%), Gaps = 4/61 (6%)
Frame = +3
Query: 424 CAICQEKMHSP----VLLRCKHIFCEECVSEWFERERTCPLCRALVKPADLQTFGDGSTS 479
CA+C + L C H F CV WF CPLCRA V+PA +Q++ + T
Sbjct: 348 CAVCLSEFTDGDECLTLPNCNHDFHSLCVEPWFASHSNCPLCRAPVQPAKVQSYTETCTV 527
Query: 480 L 480
L
Sbjct: 528 L 530
>TC88730 weakly similar to GP|21592748|gb|AAM64697.1 PRT1 {Arabidopsis
thaliana}, partial (28%)
Length = 802
Score = 46.2 bits (108), Expect = 3e-05
Identities = 15/50 (30%), Positives = 31/50 (62%)
Frame = +2
Query: 413 TMEQVNAAGDLCAICQEKMHSPVLLRCKHIFCEECVSEWFERERTCPLCR 462
T ++++ A +C +C++ + PV+L C H++CE CV + + C +C+
Sbjct: 605 TQQKISVADVMCTMCKQLLFHPVVLHCGHVYCETCVYKLADEMLRCQVCQ 754
>TC93366 similar to PIR|E96745|E96745 hypothetical protein T9N14.11
[imported] - Arabidopsis thaliana, partial (27%)
Length = 1082
Score = 45.8 bits (107), Expect = 4e-05
Identities = 22/64 (34%), Positives = 33/64 (51%), Gaps = 6/64 (9%)
Frame = +2
Query: 410 VYATME--QVNAAGDLCAICQEKMHSPVLLR----CKHIFCEECVSEWFERERTCPLCRA 463
VY+T++ ++ A CA+C + LR C H+F +CV W TCP+CRA
Sbjct: 482 VYSTVKGLKIGRAALECAVCLNEFQDDETLRLIPNCSHVFHSQCVDAWLVNHSTCPVCRA 661
Query: 464 LVKP 467
+ P
Sbjct: 662 NLIP 673
>TC77205 similar to GP|19347753|gb|AAL86301.1 putative RING-H2 zinc finger
protein ATL6 {Arabidopsis thaliana}, partial (38%)
Length = 1317
Score = 45.8 bits (107), Expect = 4e-05
Identities = 19/48 (39%), Positives = 25/48 (51%), Gaps = 4/48 (8%)
Frame = +3
Query: 424 CAICQEKMHSPVLLR----CKHIFCEECVSEWFERERTCPLCRALVKP 467
CA+C + LR C H+F EC+ EW TCP+CRA + P
Sbjct: 390 CAVCLNEFEESETLRLIPKCDHVFHPECIDEWLGSHTTCPVCRANLVP 533
>TC77204 similar to GP|21536625|gb|AAM60957.1 RING-H2 zinc finger
protein-like {Arabidopsis thaliana}, partial (30%)
Length = 1218
Score = 45.8 bits (107), Expect = 4e-05
Identities = 19/48 (39%), Positives = 25/48 (51%), Gaps = 4/48 (8%)
Frame = +3
Query: 424 CAICQEKMHSPVLLR----CKHIFCEECVSEWFERERTCPLCRALVKP 467
CA+C + LR C H+F EC+ EW TCP+CRA + P
Sbjct: 402 CAVCLMEFEDTETLRLIPKCDHVFHPECIDEWLSSHTTCPVCRANLVP 545
>AJ497196 weakly similar to SP|O60683|PEXA_ Peroxisome assembly protein 10
(Peroxin-10). [Human] {Homo sapiens}, partial (11%)
Length = 582
Score = 45.1 bits (105), Expect = 6e-05
Identities = 16/43 (37%), Positives = 22/43 (50%)
Frame = +1
Query: 424 CAICQEKMHSPVLLRCKHIFCEECVSEWFERERTCPLCRALVK 466
C++C E C H+FC C+S W + CPLCR V+
Sbjct: 310 CSLCLEARTQDTATPCGHVFCWTCISGWLQTRAQCPLCRDFVE 438
>TC79053 similar to GP|22795037|gb|AAN05420.1 putative RING protein {Populus
x canescens}, partial (28%)
Length = 1139
Score = 44.7 bits (104), Expect = 8e-05
Identities = 19/66 (28%), Positives = 34/66 (50%), Gaps = 10/66 (15%)
Frame = +2
Query: 424 CAICQEKMHSPVLLRCKHIFCEECVSEWF---------ERERTCPLCRALVKPADL-QTF 473
C IC E + PV+ C H++C C+ +W + + TCP+C+A++ L +
Sbjct: 254 CNICLESANDPVVTLCGHLYCWPCIYKWLNVQSSSVEPDTQPTCPVCKAVISHTSLVPLY 433
Query: 474 GDGSTS 479
G G ++
Sbjct: 434 GRGKSN 451
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,046,419
Number of Sequences: 36976
Number of extensions: 254169
Number of successful extensions: 1790
Number of sequences better than 10.0: 190
Number of HSP's better than 10.0 without gapping: 1727
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1764
length of query: 485
length of database: 9,014,727
effective HSP length: 100
effective length of query: 385
effective length of database: 5,317,127
effective search space: 2047093895
effective search space used: 2047093895
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0360.7


