

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
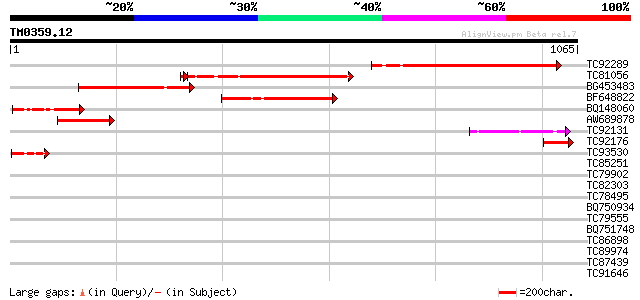
Query= TM0359.12
(1065 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC92289 weakly similar to GP|20197927|gb|AAD24632.2 hypothetical... 371 e-103
TC81056 340 2e-93
BG453483 similar to PIR|C84781|C84 hypothetical protein At2g3648... 333 3e-91
BF648822 GP|20197927|gb hypothetical protein {Arabidopsis thalia... 192 6e-49
BQ148060 homologue to GP|15450488|gb| At2g36480/F1O11.11 {Arabid... 157 2e-38
AW689878 similar to GP|4115937|gb| contains similarity to human ... 135 8e-32
TC92131 similar to GP|4115937|gb|AAD03447.1| contains similarity... 123 4e-28
TC92176 76 6e-14
TC93530 59 1e-08
TC85251 homologue to PIR|JQ2128|JQ2128 metallothionein - soybean... 41 0.002
TC79902 39 0.008
TC82303 weakly similar to GP|18073451|emb|CAC83345. GAS-2 homolo... 38 0.018
TC78495 homologue to PIR|S53504|S53504 extensin-like protein S3 ... 37 0.040
BQ750934 similar to PIR|T19673|T19 hypothetical protein C33B4.3 ... 37 0.053
TC79555 homologue to GP|10567859|gb|AAG18593.1 L8019.5 {Leishman... 37 0.053
BQ751748 similar to PIR|T47216|T47 probable V-ATPase 20K chain ... 36 0.069
TC86898 similar to GP|18376007|emb|CAB91741. probable ATP citrat... 36 0.069
TC89974 similar to GP|7767653|gb|AAF69150.1| F27F5.2 {Arabidopsi... 36 0.069
TC87439 weakly similar to GP|5139695|dbj|BAA81686.1 expressed in... 36 0.069
TC91646 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproli... 36 0.090
>TC92289 weakly similar to GP|20197927|gb|AAD24632.2 hypothetical protein
{Arabidopsis thaliana}, partial (7%)
Length = 1055
Score = 371 bits (952), Expect = e-103
Identities = 201/360 (55%), Positives = 249/360 (68%), Gaps = 4/360 (1%)
Frame = +3
Query: 680 TTTRFETAAGKGGKLSNMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSA 739
TT + K G + N IT R+LD N PS +GV+P++S G SPA LI S+
Sbjct: 3 TTKCLPAPSVKSGIIPNKSIT------RNLDASNRPSQIGVKPTRSGGPSPATLI---SS 155
Query: 740 IASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSAN-NALNPI 798
+ SLG P D S LPK+P +A + S P SSN + A+ +S++AN N LNPI
Sbjct: 156 GSPAMSLGSPDDYSPTLPKLPQGKAGKKQNDSTQPSTSSNNRGASAPSSNTANKNTLNPI 335
Query: 799 ANLLNSLVAKGLISAETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVS-GSAVPVPST 857
+NLL+SLVAKGLISA TE+ V SE + R +DQ++SI SSSLP ASV SAVPV S+
Sbjct: 336 SNLLSSLVAKGLISAGTESATTVRSETVMRSKDQTESIAVSSSLPVASVPVSSAVPVKSS 515
Query: 858 KDEVDDGAKTPISLSESTSTEIRNFIGFEFKPDVIRELHPSVISGLFDDFPHHCSICSLK 917
+ E DD AK ++LS+STSTEIRN IGF+FKPDVIRE+HP VI L D+ PHHC C ++
Sbjct: 516 RIEADDAAKASLALSQSTSTEIRNLIGFDFKPDVIREMHPHVIEELLDELPHHCGDCGIR 695
Query: 918 LKFQEQFDRHLEWHATREREHSGLIKAS-RWYLKSSDWVVGKVESLSENEFADSVDRY-G 975
LK QEQF+RHLEWHAT+ERE +GL AS RWY+ S DW+ K E LSE+EF DSVD Y
Sbjct: 696 LKQQEQFNRHLEWHATKEREQNGLTVASRRWYVTSDDWIASKAECLSESEFTDSVDEYDD 875
Query: 976 KETDRNQEDAMVLADENQCLCVLCGELFEEFYCQENGEWMFKGAVYVPNSDINDDMGIRD 1035
+TD +Q D MV+ADENQCLCVLCGELFE+ YCQE EWMFKGAVY+ N D + +M R+
Sbjct: 876 NKTDGSQLDTMVVADENQCLCVLCGELFEDVYCQERDEWMFKGAVYLNNPDSDSEMESRN 1055
>TC81056
Length = 951
Score = 340 bits (873), Expect(2) = 2e-93
Identities = 185/314 (58%), Positives = 223/314 (70%), Gaps = 2/314 (0%)
Frame = +1
Query: 334 MGTGRTGGRVAELGSDKSGYNKAAGAGVAGTISGQRNGLSIKNNFLNTEASMILDARNQP 393
MG RT GRVA+LG ++ A AGV+GTISGQRN + +K++F NTEASM +QP
Sbjct: 40 MGIVRTDGRVAKLGH--RNWSTKAAAGVSGTISGQRNSVGLKHSFSNTEASM-----HQP 198
Query: 394 RQNINSLQRGVMSNSWKHSEEEEFMWDEMNAGLTGHDAASVPNNLSKDSWTSDDENLEVE 453
+NI +QR V+S+SW++SEEEE+ WDEMN GLTGH V NNL D+WT+DDENLE E
Sbjct: 199 IRNIAGIQRNVISSSWQNSEEEEYSWDEMNTGLTGH---GVRNNLGNDAWTADDENLEAE 369
Query: 454 DNHHQSRNPFGANVDTEMSIESQATERKQLPAFRPHPSLSWKLQEQHSIDELNQKPGHSD 513
DNHHQ RN F ANVD EM SQATE+KQ AF+ HPSLSW+LQEQ SID L++KPGH +
Sbjct: 370 DNHHQIRNVFRANVDREMPNRSQATEKKQSHAFQHHPSLSWQLQEQQSIDALDRKPGHLE 549
Query: 514 GFVSTLGALPTNTNSAATRMGNRSLLPKATIGLAGNLG-QLHSVGDESPSRESSLRQQSP 572
G + G+LP N +S+A RMGNR+ LP A IGLA G Q HSVG ESPS +S LR +SP
Sbjct: 550 GSMLMSGSLPANASSSAARMGNRAFLPNARIGLAEIEGQQFHSVGSESPSGQSPLRHRSP 729
Query: 573 PPVSVHLPHLMQNLAEQDHPQTHKVSKFSGGLHSQNIKDISPALPPNIQVGKFHR-SQLR 631
P ++ HLM++LA QDHP T K S F GGLHSQ +D SP L PNI VG R SQL+
Sbjct: 730 SPPNIDHSHLMKSLAMQDHPHTRKTSHFLGGLHSQYNEDSSPTLAPNIHVGDLRRSSQLK 909
Query: 632 DLQSGSFSSMTFQP 645
DLQ G S+ FQP
Sbjct: 910 DLQ-GPLPSIGFQP 948
Score = 21.9 bits (45), Expect(2) = 2e-93
Identities = 9/14 (64%), Positives = 11/14 (78%)
Frame = +3
Query: 322 NEYGSDLSRNLGMG 335
NE+G +LSRN G G
Sbjct: 3 NEHGYNLSRNWGHG 44
>BG453483 similar to PIR|C84781|C84 hypothetical protein At2g36480 [imported]
- Arabidopsis thaliana, partial (11%)
Length = 676
Score = 333 bits (853), Expect = 3e-91
Identities = 170/219 (77%), Positives = 186/219 (84%), Gaps = 1/219 (0%)
Frame = +2
Query: 130 AIAATVCANIIEVPSEQKLPSLYLLDSIVKNIGRGYVKYFAARLPEVFCKAYRHVDPAVH 189
AIAAT+CANI+EVPSEQKLPSLYLLDSIVKNIG+ Y+KYFAA+LPEVFCKAY+ VDP+V
Sbjct: 2 AIAATICANILEVPSEQKLPSLYLLDSIVKNIGQDYIKYFAAKLPEVFCKAYKQVDPSVR 181
Query: 190 HSMRHLFGTWRGVFPSQTLQAIEKELGFTPAVNGSSSTSATLRSDSQSQRPPPSIHVNPK 249
SM+HLFGTW+GVFP QTLQ I+KELGFTP +NGSSS S LRSDSQS+ P SIHVNPK
Sbjct: 182 QSMKHLFGTWKGVFPPQTLQIIDKELGFTPVINGSSSASTALRSDSQSKHSPHSIHVNPK 361
Query: 250 YLERQRLQQSSRTKG-VDDMSGAIANSNDDPEMPDRALGVKRPRADPSNNQRAHRDAFND 308
Y ERQRLQQSSRTKG V DM+GAIANSNDD EM DR LGV R NNQR HRD FND
Sbjct: 362 YFERQRLQQSSRTKGVVGDMTGAIANSNDDHEMTDRDLGVAR----AWNNQRTHRDPFND 529
Query: 309 SVREKSISASYGGNEYGSDLSRNLGMGTGRTGGRVAELG 347
SV EKSISASY NE+GS+LSRNLGMG GRT GRVA+LG
Sbjct: 530 SVPEKSISASYEDNEHGSNLSRNLGMGIGRTDGRVAKLG 646
>BF648822 GP|20197927|gb hypothetical protein {Arabidopsis thaliana}, partial
(2%)
Length = 665
Score = 192 bits (488), Expect = 6e-49
Identities = 112/223 (50%), Positives = 146/223 (65%), Gaps = 4/223 (1%)
Frame = +2
Query: 398 NSLQRGVMSNSWKHSEEEEFMWDEMNAGLTGHDAASVPNNLSKDSWTSDDENLEVEDNHH 457
++++ MS +WK+SEEEEFMWDE+N GL+ + +V NNLS D W +DD+NLE ED H
Sbjct: 11 HNIRSSAMSKNWKNSEEEEFMWDEVNPGLSDN-VPNVSNNLSSDQWMADDDNLESED-HL 184
Query: 458 QSRNPFGANVDTEMSIESQATERKQLPAFRPHPSLSWKLQEQHSIDELNQKPGHSDGFVS 517
Q +P G V+ +S T +KQLP+ H SLSW+LQ+Q +LN KPGHS+ FVS
Sbjct: 185 QFTHPIGTKVNKGIS-----TVKKQLPSSGGHSSLSWELQKQVPSAKLNMKPGHSEIFVS 349
Query: 518 TLGALPTNTNSAATRMGNRSLLPKATIGLAGNLG--QLHSVGDESPSRESS-LRQQSPP- 573
LP N NS+A R+ N+S +P TIG++ G Q S G ESPS +SS LRQQSP
Sbjct: 350 APSGLPKNPNSSAARIRNQSSMPHTTIGMSKITGQQQFDSEGTESPSEQSSPLRQQSPKV 529
Query: 574 PVSVHLPHLMQNLAEQDHPQTHKVSKFSGGLHSQNIKDISPAL 616
PV++ P M+NLAEQD P T K S+ GGL SQ I+D PA+
Sbjct: 530 PVTIRNPPSMRNLAEQDCPTTLKTSQHLGGLQSQYIRDPVPAI 658
>BQ148060 homologue to GP|15450488|gb| At2g36480/F1O11.11 {Arabidopsis
thaliana}, partial (25%)
Length = 667
Score = 157 bits (398), Expect = 2e-38
Identities = 89/134 (66%), Positives = 105/134 (77%)
Frame = +1
Query: 6 RRPFDRSREPGLKRPRMMEEPERVAAAQNPSSRQFLPQRQQQVAASGISTLSSAARLRAA 65
RRPFDRS+EP LKRPR++E P+R A N S R P +Q++ SGISTLSSA+R R
Sbjct: 301 RRPFDRSKEPILKRPRLIENPDR---APNSSVR---PIQQRKQVNSGISTLSSASRFRQ- 459
Query: 66 ANSRDLESSDFGRGDGGGGYHPRPPPFQELVAQYKSALAELTFNSKPIITNLTIIAGENQ 125
N R+LESSDF GGGY P+P F++LVAQYKSALAELTFNSKP+ITNLTIIAGENQ
Sbjct: 460 -NDRELESSDFD----GGGYEPQPIRFEDLVAQYKSALAELTFNSKPMITNLTIIAGENQ 624
Query: 126 AAEQAIAATVCANI 139
A ++AIAAT+CANI
Sbjct: 625 AGQKAIAATICANI 666
>AW689878 similar to GP|4115937|gb| contains similarity to human PCF11p
homolog (GB:AF046935) {Arabidopsis thaliana}, partial
(14%)
Length = 635
Score = 135 bits (340), Expect = 8e-32
Identities = 65/108 (60%), Positives = 85/108 (78%)
Frame = +2
Query: 90 PPFQELVAQYKSALAELTFNSKPIITNLTIIAGENQAAEQAIAATVCANIIEVPSEQKLP 149
P +E+V Y+ L+ELT N KPIIT+LTIIA +++ + IA +C I+EV ++QKLP
Sbjct: 275 PSTEEIVQIYELLLSELTCNVKPIITDLTIIAEQHRNHAKGIAFAICNRIVEVSADQKLP 454
Query: 150 SLYLLDSIVKNIGRGYVKYFAARLPEVFCKAYRHVDPAVHHSMRHLFG 197
SLYLLDSIVKN+G+ YV+YF+ RLPEVFC AYR VDP++H +MRHLFG
Sbjct: 455 SLYLLDSIVKNVGQEYVRYFSQRLPEVFCVAYRQVDPSLHSAMRHLFG 598
>TC92131 similar to GP|4115937|gb|AAD03447.1| contains similarity to human
PCF11p homolog (GB:AF046935) {Arabidopsis thaliana},
partial (24%)
Length = 1003
Score = 123 bits (308), Expect = 4e-28
Identities = 67/192 (34%), Positives = 102/192 (52%), Gaps = 4/192 (2%)
Frame = +3
Query: 865 AKTPISLSESTSTEIRNFIGFEFKPDVIRELHPSVISGLFDDFPHHCSICSLKLKFQEQF 924
A+ ISL+ ++ +F+G EF PD ++ H S IS L+ + P C+ C L+ K Q++
Sbjct: 384 AQGVISLTNQAPSQ--DFVGIEFDPDTLKVRHESAISALYGNLPRQCTTCGLRFKSQDEH 557
Query: 925 DRHLEWHATRER--EHSGLIKASRWYLKSSDWVVGKVESLSENEFADSVDRYGKETDRNQ 982
H++WH T+ R ++ + +W++ + W+ G E+L + E +
Sbjct: 558 SSHMDWHVTKNRMSKNRKQKPSRKWFVSETMWLXG-AEALGTESAPGFLPTETTEEKKED 734
Query: 983 EDAMVLADENQCLCVLCGELFEEFYCQENGEWMFKGAVYVPNSDINDDMGIRDASTGG-- 1040
E+ V ADE+Q C LCGE FEEFY E EWM++GAVY +N GI
Sbjct: 735 EELAVPADEDQNTCALCGEPFEEFYSDETEEWMYRGAVY-----LNAPNGITTGMDRSXL 899
Query: 1041 GPIIHARCLSEN 1052
G IIHA+C SE+
Sbjct: 900 GSIIHAKCXSES 935
>TC92176
Length = 757
Score = 76.3 bits (186), Expect = 6e-14
Identities = 33/56 (58%), Positives = 42/56 (74%)
Frame = +1
Query: 1003 FEEFYCQENGEWMFKGAVYVPNSDINDDMGIRDASTGGGPIIHARCLSENPVSSVL 1058
FE+ YC EN EWMFK AVY+ N+DI ++GI D +G GPIIH RCLS+ +SSV+
Sbjct: 1 FEDVYCHENSEWMFKAAVYMTNADIATEVGINDVISGRGPIIHTRCLSDYSLSSVV 168
>TC93530
Length = 443
Score = 58.9 bits (141), Expect = 1e-08
Identities = 36/72 (50%), Positives = 48/72 (66%)
Frame = +3
Query: 4 SSRRPFDRSREPGLKRPRMMEEPERVAAAQNPSSRQFLPQRQQQVAASGISTLSSAARLR 63
+SRR DRSREPG K+PR+++E ++ N +SR F PQRQQ SG++T+ S+ R R
Sbjct: 243 NSRRSLDRSREPGAKKPRLIDELQQ---GSNQTSRTF-PQRQQ--PTSGVATMLSSGRFR 404
Query: 64 AAANSRDLESSD 75
N RD ESSD
Sbjct: 405 --MNDRDSESSD 434
>TC85251 homologue to PIR|JQ2128|JQ2128 metallothionein - soybean, partial
(93%)
Length = 1554
Score = 41.2 bits (95), Expect = 0.002
Identities = 38/148 (25%), Positives = 59/148 (39%), Gaps = 3/148 (2%)
Frame = -2
Query: 653 SSHTEVTAKTEKPHLSKVPLPRETEQPTTTRFETAAGKGGKLSNMPITNRLPTSRSLDTG 712
++ TA T+ P + + T QP T +++ PIT++ P +
Sbjct: 860 NAQAPATAPTKLPPTTPTAITPVTTQPPTV-----------VASPPITSQPPVT------ 732
Query: 713 NLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQP---PR 769
V P + +SPA I P S+ PP PK + + P +PP P P
Sbjct: 731 -------VAPKSAPVTSPAPKIAPASSPKVPPP-QPPKSSPVSTPTLPPPLPPPPKISPT 576
Query: 770 TSALPPASSNLKSAAVQASSSANNALNP 797
PPA + +K+ V A + A A P
Sbjct: 575 PVQTPPAPAPVKATPVPAPAPAKQAPTP 492
>TC79902
Length = 518
Score = 39.3 bits (90), Expect = 0.008
Identities = 27/92 (29%), Positives = 41/92 (44%)
Frame = +3
Query: 696 NMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSA 755
NM I + T+ L T + +QS SSP PP AI+ P+ P + S+
Sbjct: 81 NMNIALMIVTALLLSTTTI--------AQSPASSPTKSQPPRKAISPSPAASSPPEPSAT 236
Query: 756 LPKIPPRRAAQPPRTSALPPASSNLKSAAVQA 787
P + P + PP + P +S+ L +V A
Sbjct: 237 SPAVSPSSISGPPSEAPGPASSAVLNRVSVAA 332
>TC82303 weakly similar to GP|18073451|emb|CAC83345. GAS-2 homologue
{Candida glabrata}, partial (7%)
Length = 846
Score = 38.1 bits (87), Expect = 0.018
Identities = 41/133 (30%), Positives = 56/133 (41%), Gaps = 4/133 (3%)
Frame = +1
Query: 673 PRETEQPTTTRFETAAGKGGKLSNMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAG 732
P PT TA +S T+ PT+R T P P +S + P
Sbjct: 130 PAPVSAPTPPLVSTALTACATMSRSLPTSWTPTTR---TRRRPP-----PPVTSTARPRP 285
Query: 733 LIPPVSAIASPPSLGRPKDNSS-ALPKIPPR-RAAQPPRTSALP--PASSNLKSAAVQAS 788
PP +A+ASP S P +S P +PP AA PP A P ASS +S+ + AS
Sbjct: 286 APPPATALASPLSPPLPPPTASLPPPPVPPMPLAALPPALPATPLLVASSPSRSSPLAAS 465
Query: 789 SSANNALNPIANL 801
S P+ ++
Sbjct: 466 PSVPTLSLPVLSV 504
>TC78495 homologue to PIR|S53504|S53504 extensin-like protein S3 - alfalfa,
partial (97%)
Length = 1049
Score = 37.0 bits (84), Expect = 0.040
Identities = 22/57 (38%), Positives = 28/57 (48%)
Frame = +3
Query: 722 PSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALPPASS 778
PS S SSPA PP + + P P SS PP +++ PP S+ PPA S
Sbjct: 243 PSTSPNSSPAPPTPPANTPPTTPQASPPPVQSSP----PPVQSSPPPLQSSPPPAQS 401
>BQ750934 similar to PIR|T19673|T19 hypothetical protein C33B4.3 -
Caenorhabditis elegans, partial (1%)
Length = 627
Score = 36.6 bits (83), Expect = 0.053
Identities = 39/164 (23%), Positives = 67/164 (40%), Gaps = 8/164 (4%)
Frame = +2
Query: 722 PSQSSGSSPAGLIPPVSAIASPPSLGRPKDN----SSALPKIPPRRAAQPPRTSALPPAS 777
P G+ G + P +D+ ++A + AA P TSA+ +
Sbjct: 68 PCAGDGTQKCGAGDRLQVYLFSPEASATEDSVLTSTTASSSVETTAAAVFPTTSAVANGT 247
Query: 778 SNLKSAAVQASSSANNALNPIANLLNSLVAKGLIS--AETETTAKVPSEMLTRLE--DQS 833
+A+ + ++ + NS V S AE ETT+ V S T LE D +
Sbjct: 248 LTAVTASETVTDEVASSSLSTLSATNSSVTSAAPSFTAEAETTSPVSSASSTSLETLDAT 427
Query: 834 DSITTSSSLPGASVSGSAVPVPSTKDEVDDGAKTPISLSESTST 877
+ SSS+P + S +++P A +P S++ ST++
Sbjct: 428 TTAVPSSSIPES--SSTSLPPTLVPQNTTSPASSPSSITTSTTS 553
>TC79555 homologue to GP|10567859|gb|AAG18593.1 L8019.5 {Leishmania major},
partial (1%)
Length = 1410
Score = 36.6 bits (83), Expect = 0.053
Identities = 28/92 (30%), Positives = 41/92 (44%)
Frame = +2
Query: 700 TNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKI 759
+N P+S +G LP+ P+ ++ S P PP S +S P P S+ P
Sbjct: 1025 SNTCPSSPRRRSGRLPAATTSSPATNANSPP---FPPASPSSSVPPAKPPTSKSARSPAP 1195
Query: 760 PPRRAAQPPRTSALPPASSNLKSAAVQASSSA 791
R + PP +S P S+ S+ ASS A
Sbjct: 1196 V*SRPSPPPSSS---PGSAASASSRCSASSRA 1282
Score = 34.3 bits (77), Expect = 0.26
Identities = 38/146 (26%), Positives = 59/146 (40%), Gaps = 8/146 (5%)
Frame = +2
Query: 672 LPRETEQPTTTRFETAAGKGGKLSNMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPA 731
+P + QP++TR A + S+ T R P + PS S SS +P+
Sbjct: 467 VPP*SPQPSSTRSSPAPPS--RPSSPATTTRPPPTSKPSPTQTPS-----RSPSSARTPS 625
Query: 732 GLIPPVSA--IASPPS----LGRPKDNSSALPKIP--PRRAAQPPRTSALPPASSNLKSA 783
P A ++SPP + + +LPK P PR A P A P L +A
Sbjct: 626 TSSTPTRAPLLSSPPPPPPFFPARRPGTRSLPKSPPRPRPARLPSARRASPRGGRPLAAA 805
Query: 784 AVQASSSANNALNPIANLLNSLVAKG 809
A ++++ A P + S +G
Sbjct: 806 AAAGATTSTTASRPTRSARTSAARRG 883
>BQ751748 similar to PIR|T47216|T47 probable V-ATPase 20K chain [imported] -
Neurospora crassa, partial (82%)
Length = 642
Score = 36.2 bits (82), Expect = 0.069
Identities = 36/147 (24%), Positives = 59/147 (39%), Gaps = 5/147 (3%)
Frame = -1
Query: 735 PPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSANNA 794
PP ++ P+ S +P R + PPR PPAS+ + + SSS +
Sbjct: 639 PPAPTLSPSPATTSASATS*CSRAVPARSSRSPPRP---PPASTAISVSTSSPSSSTRSP 469
Query: 795 LNPIANLLNSLVAKGLISAETETTAKVPSEMLTRLEDQSDSITTSSSLPG--ASVSGSAV 852
P + L + SA + ++ + + L +T+S + P + SG A
Sbjct: 468 --PSSPTLPPASSSRPCSAPSSSSIVSSTSPMVTLSP*PRPVTSSRACPSLTSPTSGPAS 295
Query: 853 PVPSTKDEVDDGAK---TPISLSESTS 876
PST + TP++ S STS
Sbjct: 294 TAPSTPAVAASASSS*TTPVASSPSTS 214
Score = 35.8 bits (81), Expect = 0.090
Identities = 36/139 (25%), Positives = 56/139 (39%), Gaps = 10/139 (7%)
Frame = -1
Query: 723 SQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALP---KIPPRRAAQPPRTSALPPASSN 779
+ +S +S P + SPP RP S+A+ P PP + LPPASS+
Sbjct: 603 TSASATS*CSRAVPARSSRSPP---RPPPASTAISVSTSSPSSSTRSPPSSPTLPPASSS 433
Query: 780 LKSAAVQASSSANNALNPIANL-------LNSLVAKGLISAETETTAKVPSEMLTRLEDQ 832
+ + +SSS ++ +P+ L +S L S + + PS
Sbjct: 432 -RPCSAPSSSSIVSSTSPMVTLSP*PRPVTSSRACPSLTSPTSGPASTAPSTPAVAASAS 256
Query: 833 SDSITTSSSLPGASVSGSA 851
S T +S P S S +A
Sbjct: 255 SS*TTPVASSPSTSRSSTA 199
>TC86898 similar to GP|18376007|emb|CAB91741. probable ATP citrate lyase
subunit 2 {Neurospora crassa}, complete
Length = 2389
Score = 36.2 bits (82), Expect = 0.069
Identities = 59/243 (24%), Positives = 92/243 (37%), Gaps = 23/243 (9%)
Frame = +3
Query: 639 SSMTFQPSHKQQLGSSHTEVTAKTEKPHLSKVPLPRETEQPTTTRFETAAGKGGKLSNMP 698
S+ T + SS + + P S + LP T PT + A G L +P
Sbjct: 1212 STSTASSPTSRSTPSSSSPTRTLPQPPCTSWILLPSSTRPPTLS-----AVSSGPLPALP 1376
Query: 699 ITNRLPT-------------SRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPS 745
+ + PT +R + L + R +S + +PP S ++S P+
Sbjct: 1377 LPSA*PTLPLPTAKRSTSMRARRWSSPLLSVVS*PRRRLTSPTLMPRPVPPSSLLSSTPT 1556
Query: 746 ----LGRP---KDNSSALPKIPPRRAAQPPRTSA---LPPASSNLKSAAVQASSSANNAL 795
L P +S+ P PP P T + LPP S + V +S+S + L
Sbjct: 1557 AVSGLSSPVVVPPSSTPTPSPPPASPTSSPTTVSTLVLPP-SPRPTTTPVLSSTSCSALL 1733
Query: 796 NPIANLLNSLVAKGLISAETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVSGSAVPVP 855
P A+ +S T+ S +R+ + T SSS SGS VP P
Sbjct: 1734 *PRR-------ARSSLSVVVLPTSPT-SPAHSRVSSGPSATTLSSSTSTRPKSGSVVPAP 1889
Query: 856 STK 858
+T+
Sbjct: 1890 TTR 1898
>TC89974 similar to GP|7767653|gb|AAF69150.1| F27F5.2 {Arabidopsis
thaliana}, partial (12%)
Length = 1479
Score = 36.2 bits (82), Expect = 0.069
Identities = 27/81 (33%), Positives = 44/81 (53%), Gaps = 10/81 (12%)
Frame = +2
Query: 730 PAGLIPPVSAIASPPSLGRPKDNSSALPKI--PPRRAAQP---PRTSALP---PASSNLK 781
P PP S PS G+ +DN++ALP+ PP+ A P P S++P A ++++
Sbjct: 887 PVSGAPPQSLYTFTPSYGQQQDNANALPQYQHPPQMHAPPAGQPWLSSVPKSAAAVTSVQ 1066
Query: 782 SAAVQASSSANN--ALNPIAN 800
+ VQ+S +A+ A N +N
Sbjct: 1067PSGVQSSGTASTDAATNTTSN 1129
>TC87439 weakly similar to GP|5139695|dbj|BAA81686.1 expressed in cucumber
hypocotyls {Cucumis sativus}, partial (57%)
Length = 897
Score = 36.2 bits (82), Expect = 0.069
Identities = 28/91 (30%), Positives = 41/91 (44%), Gaps = 16/91 (17%)
Frame = +3
Query: 725 SSGSSPAGLIPPVSAIASPPS-------LGRPKDN------SSALPKIPPRRAAQPPR-- 769
+S S+PA P S +A+PP+ + PK + +A P P A+ PP
Sbjct: 126 TSPSTPAATTPVSSPVAAPPTTPTTPAPVASPKSSPPATSPKAAAPTATPPAASSPPAVT 305
Query: 770 -TSALPPASSNLKSAAVQASSSANNALNPIA 799
S PPA +KS A S+ A+ P+A
Sbjct: 306 PVSTPPPAPVPVKSPPTPAPVSSPPAVTPVA 398
>TC91646 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproline-rich
glycoprotein DZ-HRGP {Volvox carteri f. nagariensis},
partial (15%)
Length = 984
Score = 35.8 bits (81), Expect = 0.090
Identities = 58/224 (25%), Positives = 78/224 (33%), Gaps = 8/224 (3%)
Frame = +1
Query: 662 TEKPHLSKVPLPRETEQPTTTRFETAAGKGGKLSNMPITNRLPTSRSLDTGNLPSILGVR 721
T P L+ P P + P + +L + P LP S S N P +
Sbjct: 313 TTPPSLTTSPSPTASPPPRSPPRPPPRSPPRQLMSPP--RPLPRSPSPPPRNPPELSRTT 486
Query: 722 PS-------QSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALP 774
P+ S +SP PPVS P+L R P +P A+ S LP
Sbjct: 487 PTPRPPMSPSPSPTSPPAATPPVSRPPPRPALRRLLPPPPLPPTLPSSMASLFLAASVLP 666
Query: 775 PASSNLKSAAVQASSSANNALNPIANLLNSLVAKGLISAETETTAKVPS-EMLTRLEDQS 833
PAS SS A LL L A + +T + V S TR
Sbjct: 667 PAS------PPTT*SSPPTP*PSSAALLLCLPASATLPCSEQTASPVTSWTTSTRCPSAR 828
Query: 834 DSITTSSSLPGASVSGSAVPVPSTKDEVDDGAKTPISLSESTST 877
S T S+ + ++ +AV V V + S S S ST
Sbjct: 829 TSATLSAPV---TLPSAAVVVLPLLSSVSSSPRPSASPSTSAST 951
Score = 30.4 bits (67), Expect = 3.8
Identities = 18/55 (32%), Positives = 25/55 (44%)
Frame = +1
Query: 722 PSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALPPA 776
PS+ ++P L S ASPP P+ P+ PPR+ PPR P+
Sbjct: 295 PSRLPLTTPPSLTTSPSPTASPPPRSPPRPP----PRSPPRQLMSPPRPLPRSPS 447
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.311 0.128 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,812,448
Number of Sequences: 36976
Number of extensions: 452645
Number of successful extensions: 3160
Number of sequences better than 10.0: 128
Number of HSP's better than 10.0 without gapping: 2830
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3066
length of query: 1065
length of database: 9,014,727
effective HSP length: 106
effective length of query: 959
effective length of database: 5,095,271
effective search space: 4886364889
effective search space used: 4886364889
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0359.12


