

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
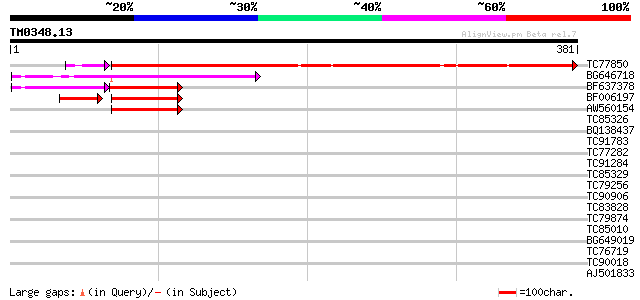
Query= TM0348.13
(381 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77850 weakly similar to PIR|H96517|H96517 protein T2E6.22 [imp... 311 2e-86
BG646718 weakly similar to PIR|H96517|H96 protein T2E6.22 [impor... 100 1e-21
BF637378 weakly similar to PIR|H96517|H965 protein T2E6.22 [impo... 47 2e-11
BF006197 weakly similar to GP|1310677|emb| protein z-type serpin... 40 1e-06
AW560154 42 3e-04
TC85326 similar to GP|5738542|emb|CAB53034.1 photosystem I subun... 31 0.69
BQ138437 weakly similar to PIR|T51919|T519 related to aldehyde d... 30 1.2
TC91783 similar to GP|16226796|gb|AAL16264.1 At2g47960/T9J23.10 ... 30 1.5
TC77282 homologue to GP|17065300|gb|AAL32804.1 Unknown protein {... 29 2.6
TC91284 similar to GP|21554350|gb|AAM63457.1 unknown {Arabidopsi... 29 2.6
TC85329 homologue to GP|5738542|emb|CAB53034.1 photosystem I sub... 29 2.6
TC79256 similar to GP|21554235|gb|AAM63310.1 CPRD49 {Arabidopsis... 29 3.4
TC90906 similar to GP|16604607|gb|AAL24096.1 putative AtBgamma p... 28 4.5
TC83828 weakly similar to GP|22530894|gb|AAM96888.1 hypothetical... 28 4.5
TC79874 similar to GP|17065608|gb|AAL33784.1 unknown protein {Ar... 28 4.5
TC85010 weakly similar to PIR|T40514|T40514 Chaperonin hsp78p - ... 28 4.5
BG649019 similar to PIR|T52612|T52 SRP receptor homolog FtsY pre... 28 5.9
TC76719 similar to GP|21595178|gb|AAM66078.1 endo-xyloglucan tra... 28 5.9
TC90018 weakly similar to PIR|B84607|B84607 hypothetical protein... 28 7.7
AJ501833 PIR|A87356|A87 sugar ABC transporter ATP-binding prote... 28 7.7
>TC77850 weakly similar to PIR|H96517|H96517 protein T2E6.22 [imported] -
Arabidopsis thaliana, partial (65%)
Length = 1485
Score = 311 bits (798), Expect(2) = 2e-86
Identities = 172/314 (54%), Positives = 228/314 (71%), Gaps = 1/314 (0%)
Frame = +2
Query: 69 DAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLI 128
DA+ A LSF +G++VD+++SL F+ +++ + A+L+S+DF+N+ V ++VNS
Sbjct: 284 DASPAGGPLLSFVDGVWVDQTLSLQPSFQQIVSTHFKAALSSVDFQNKAVEVTNEVNSWA 463
Query: 129 ELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKFDGPRYE-FPFDLLNGTSVKVP 187
E ++NG I +LLP G++ N TRLIFANAL F+G W KFD + E + F LLNG+ VKVP
Sbjct: 464 EKETNGLIKELLPLGSVNNATRLIFANALYFKGAWNDKFDASKTEDYEFHLLNGSPVKVP 643
Query: 188 FMTIKKKTHYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPGF 247
FMT KKK +IRAFD FK+L LPYKQG D KR+F+M FLP+A DGL+ALV K++SE
Sbjct: 644 FMTSKKK-QFIRAFDGFKVLGLPYKQGED-KRQFTMYFFLPNAKDGLAALVEKVASESEL 817
Query: 248 LKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSNN 307
L+ KLP KV V +IPKFNISF E S++LKE+GVV PFS TKM+ +S S N
Sbjct: 818 LQHKLPFGKVEVGDFRIPKFNISFGLETSDMLKELGVVLPFS--GGGLTKMVN-SSVSQN 988
Query: 308 LCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIANHPFLFLIREDFTG 367
LCV +IFHK+FIEVNE+GT+A AA+ + +SA P ++F+A+HPFLF+IRED TG
Sbjct: 989 LCVSNIFHKSFIEVNEEGTEAAAATAA-TILLRSAMSIPPRLDFVADHPFLFMIREDLTG 1165
Query: 368 TILFVGQVLNPLDG 381
TI+FVGQVLNPL G
Sbjct: 1166TIIFVGQVLNPLAG 1207
Score = 25.4 bits (54), Expect(2) = 2e-86
Identities = 15/30 (50%), Positives = 17/30 (56%)
Frame = +3
Query: 38 STSFSPSSDLTPSTISTPSSLRGFPLCSPT 67
STSF+P+ P IST S FP SPT
Sbjct: 210 STSFNPN----PPIISTTSLHSSFPSSSPT 287
>BG646718 weakly similar to PIR|H96517|H96 protein T2E6.22 [imported] -
Arabidopsis thaliana, partial (30%)
Length = 702
Score = 100 bits (248), Expect = 1e-21
Identities = 64/169 (37%), Positives = 95/169 (55%), Gaps = 2/169 (1%)
Frame = +2
Query: 2 NIYSQKKIIKRRTSSIHPCLSTLLLASWPPVQKAAHSTSFSPSSDLTPSTISTPSSLRGF 61
NI SQ ++ TSS CL L A +H+ + +S L I+ S L
Sbjct: 62 NICSQNN--QKTTSSFRHCLCKLCSAF*-----LSHTAA---TSRLPTIQIN*SSQLPRL 211
Query: 62 PLCSPT--KDAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDR 119
C DAA A HLSF +G++++K++SL F+ +++ Y A+LAS+DF +
Sbjct: 212 TACFSIVLSDAASAGGPHLSFVDGVWIEKTLSLQPSFKQIMSSDYKATLASVDFMFKAPE 391
Query: 120 VLHDVNSLIELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKFD 168
V +VN +E ++NG I +L+PPG++ +LT LIFANAL F+G W KFD
Sbjct: 392 VRKEVNIWVEKETNGLIKELIPPGSVDSLTSLIFANALYFRGAWNEKFD 538
>BF637378 weakly similar to PIR|H96517|H965 protein T2E6.22 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 627
Score = 47.0 bits (110), Expect(2) = 2e-11
Identities = 19/49 (38%), Positives = 35/49 (70%)
Frame = +1
Query: 68 KDAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNR 116
KDA + HLSF +G++++K++SL F+ +++ Y A+L+ +DFKN+
Sbjct: 247 KDATSSGGPHLSFVDGVWLEKTLSLQSSFKQIVSEDYKATLSEVDFKNK 393
Score = 39.3 bits (90), Expect(2) = 2e-11
Identities = 28/66 (42%), Positives = 32/66 (48%)
Frame = +2
Query: 2 NIYSQKKIIKRRTSSIHPCLSTLLLASWPPVQKAAHSTSFSPSSDLTPSTISTPSSLRGF 61
NI S K ++RTS CLS L A + H+ SF SS P ISTP SL F
Sbjct: 62 NISSPKN--QKRTSCFRHCLSKLCSA*SLLALRVQHNNSFLTSSVSNPLIISTPQSLINF 235
Query: 62 PLCSPT 67
P S T
Sbjct: 236 PSSSKT 253
>BF006197 weakly similar to GP|1310677|emb| protein z-type serpin {Hordeum
vulgare subsp. vulgare}, partial (7%)
Length = 468
Score = 40.0 bits (92), Expect(2) = 1e-06
Identities = 19/48 (39%), Positives = 31/48 (64%)
Frame = +3
Query: 69 DAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNR 116
DAA + LS T ++VD+++SL F+ + Y A+LAS+DF+N+
Sbjct: 120 DAAPSGGSRLSLTQRVWVDQTLSLQPSFKETMVTDYKATLASVDFQNK 263
Score = 30.0 bits (66), Expect(2) = 1e-06
Identities = 15/29 (51%), Positives = 18/29 (61%)
Frame = +1
Query: 34 KAAHSTSFSPSSDLTPSTISTPSSLRGFP 62
+ H+ +FS SS STISTPS L FP
Sbjct: 22 RVVHNNNFSNSSVPNSSTISTPSLLISFP 108
>AW560154
Length = 470
Score = 42.4 bits (98), Expect = 3e-04
Identities = 20/48 (41%), Positives = 32/48 (66%)
Frame = +1
Query: 69 DAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNR 116
DAA + LSFT ++VD+++SL F+ + Y A+LAS+DF+N+
Sbjct: 49 DAAPSGGSRLSFTQRVWVDQTLSLQPSFKETMVTDYKATLASVDFQNK 192
>TC85326 similar to GP|5738542|emb|CAB53034.1 photosystem I subunit XI
precursor {Arabidopsis thaliana}, partial (85%)
Length = 826
Score = 31.2 bits (69), Expect = 0.69
Identities = 32/106 (30%), Positives = 51/106 (47%), Gaps = 3/106 (2%)
Frame = +2
Query: 96 FRLLLAYQYNASLASLDFKNRGDRVL---HDVNSLIELQSNGHITQLLPPGTITNLTRLI 152
++LLL +Q N+S ASLD + + + H +N L+E G++T LL P + + T
Sbjct: 47 WQLLLLWQANSSPASLDLWSHLEDFVALPHSINYLLE----GNLTSLLRPSNLKSQTTK* 214
Query: 153 FANALRFQGLWKHKFDGPRYEFPFDLLNGTSVKVPFMTIKKKTHYI 198
F ++ L K + LL+GTS T + THY+
Sbjct: 215 FNQSMVIHSLEALKLQLHQAH----LLHGTSPTYQ-ATELQ*THYL 337
>BQ138437 weakly similar to PIR|T51919|T519 related to aldehyde dehydrogenase
(NAD+) [imported] - Neurospora crassa, partial (14%)
Length = 668
Score = 30.4 bits (67), Expect = 1.2
Identities = 16/45 (35%), Positives = 26/45 (57%), Gaps = 2/45 (4%)
Frame = -1
Query: 116 RGDRVLHDVNSLIEL--QSNGHITQLLPPGTITNLTRLIFANALR 158
RG+ +L NS++++ NGH+ QLL G + + L+ A A R
Sbjct: 206 RGEGLLCSCNSIVQVLVARNGHVPQLLAGGRVDAMVDLVRATASR 72
>TC91783 similar to GP|16226796|gb|AAL16264.1 At2g47960/T9J23.10
{Arabidopsis thaliana}, partial (27%)
Length = 597
Score = 30.0 bits (66), Expect = 1.5
Identities = 18/61 (29%), Positives = 27/61 (43%)
Frame = +1
Query: 3 IYSQKKIIKRRTSSIHPCLSTLLLASWPPVQKAAHSTSFSPSSDLTPSTISTPSSLRGFP 62
I+S + S +HP STLL S P + +TS + P + P+SL P
Sbjct: 142 IFSLSSLSCHADSVVHPSTSTLLFVSIPTTSSSVKTTS------MIPPPLPPPTSLLPIP 303
Query: 63 L 63
+
Sbjct: 304 I 306
>TC77282 homologue to GP|17065300|gb|AAL32804.1 Unknown protein {Arabidopsis
thaliana}, partial (19%)
Length = 787
Score = 29.3 bits (64), Expect = 2.6
Identities = 19/57 (33%), Positives = 26/57 (45%), Gaps = 5/57 (8%)
Frame = +2
Query: 31 PVQKAAHSTSFSPSSDLTPSTISTPSSLRGFPL-----CSPTKDAADADAHHLSFTN 82
P + STS SP S +P+T S P+ P +P + ADA + S TN
Sbjct: 269 PSPAVSPSTSSSPGSSPSPNTFSPPAPGSNVPASGPGGATPGQPGADAPSAAFSITN 439
>TC91284 similar to GP|21554350|gb|AAM63457.1 unknown {Arabidopsis
thaliana}, partial (77%)
Length = 742
Score = 29.3 bits (64), Expect = 2.6
Identities = 18/55 (32%), Positives = 29/55 (52%)
Frame = +1
Query: 31 PVQKAAHSTSFSPSSDLTPSTISTPSSLRGFPLCSPTKDAADADAHHLSFTNGMF 85
P +A+S++ SPS+ TP T+STP+ P SP + +H +S + F
Sbjct: 67 PSSPSAYSSA-SPSTQTTPPTLSTPT-----PPASPPPPSPKISSHFMSTHSAHF 213
>TC85329 homologue to GP|5738542|emb|CAB53034.1 photosystem I subunit XI
precursor {Arabidopsis thaliana}, partial (79%)
Length = 1068
Score = 29.3 bits (64), Expect = 2.6
Identities = 19/53 (35%), Positives = 31/53 (57%), Gaps = 3/53 (5%)
Frame = +1
Query: 96 FRLLLAYQYNASLASLDFKNRGDRVL---HDVNSLIELQSNGHITQLLPPGTI 145
++LLL +Q N+S ASLD + + + H +N L+E G++T LL P +
Sbjct: 16 WQLLLLWQANSSPASLDLWSHLEDFVALPHSINYLLE----GNLTSLLRPSNL 162
>TC79256 similar to GP|21554235|gb|AAM63310.1 CPRD49 {Arabidopsis thaliana},
complete
Length = 1261
Score = 28.9 bits (63), Expect = 3.4
Identities = 21/50 (42%), Positives = 25/50 (50%), Gaps = 3/50 (6%)
Frame = -3
Query: 15 SSIHPCLSTLLL---ASWPPVQKAAHSTSFSPSSDLTPSTISTPSSLRGF 61
S+ + +T+LL A W P K A S S S L ST TPSSL F
Sbjct: 815 STFNISFTTILLPSAAKWIPSVKHAFSQSSILCSALKRSTTFTPSSLHSF 666
>TC90906 similar to GP|16604607|gb|AAL24096.1 putative AtBgamma protein
{Arabidopsis thaliana}, partial (12%)
Length = 863
Score = 28.5 bits (62), Expect = 4.5
Identities = 16/64 (25%), Positives = 29/64 (45%)
Frame = +2
Query: 135 HITQLLPPGTITNLTRLIFANALRFQGLWKHKFDGPRYEFPFDLLNGTSVKVPFMTIKKK 194
H +L P + + + F +L F L+ H + F F +L + + +PF+ I
Sbjct: 101 HFPKLYPNHKTSLICIITFLFSL*FSFLFSHSREKKSENFDFHMLI*SIIILPFLPITHN 280
Query: 195 THYI 198
HY+
Sbjct: 281 LHYL 292
>TC83828 weakly similar to GP|22530894|gb|AAM96888.1 hypothetical protein
{Arabidopsis thaliana}, partial (20%)
Length = 700
Score = 28.5 bits (62), Expect = 4.5
Identities = 21/77 (27%), Positives = 38/77 (49%), Gaps = 5/77 (6%)
Frame = -2
Query: 238 VHKLSSEPGFLKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEVG-----VVSPFSPMD 292
VH++S E F K P+RK + + + + +S+ LK+ G + SP++
Sbjct: 597 VHRMSKENRFKIYKQPKRKEQYTSKLQCVSSFAHFASSSDALKDSGFNILFTETSISPLE 418
Query: 293 ACFTKMLEINSPSNNLC 309
A F++ + + SNN C
Sbjct: 417 AAFSE-IHSGTNSNNKC 370
>TC79874 similar to GP|17065608|gb|AAL33784.1 unknown protein {Arabidopsis
thaliana}, partial (88%)
Length = 1154
Score = 28.5 bits (62), Expect = 4.5
Identities = 18/40 (45%), Positives = 22/40 (55%)
Frame = +1
Query: 30 PPVQKAAHSTSFSPSSDLTPSTISTPSSLRGFPLCSPTKD 69
PP + TSFSPSS +TP T + PSS P SP +
Sbjct: 298 PPTPSHLNLTSFSPSS-VTPPTFA-PSSSTQTPALSPVSN 411
>TC85010 weakly similar to PIR|T40514|T40514 Chaperonin hsp78p - fission
yeast (Schizosaccharomyces pombe), partial (23%)
Length = 904
Score = 28.5 bits (62), Expect = 4.5
Identities = 22/61 (36%), Positives = 30/61 (49%), Gaps = 2/61 (3%)
Frame = +1
Query: 1 HNIYSQKKIIKRRT--SSIHPCLSTLLLASWPPVQKAAHSTSFSPSSDLTPSTISTPSSL 58
H YS +RRT S + C S + + S ++A STS PSS P++ T SS
Sbjct: 97 HTRYSSSTSSRRRTATSRLSSCRSLMRVTS--RTRRATRSTSAIPSSYSHPTSAPTFSSA 270
Query: 59 R 59
R
Sbjct: 271 R 273
>BG649019 similar to PIR|T52612|T52 SRP receptor homolog FtsY precursor
chloroplast [validated] - Arabidopsis thaliana, partial
(39%)
Length = 788
Score = 28.1 bits (61), Expect = 5.9
Identities = 22/65 (33%), Positives = 33/65 (49%)
Frame = -1
Query: 46 DLTPSTISTPSSLRGFPLCSPTKDAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYN 105
D++ +TIS PSS F C T DA D F D + ++S+P +L A+ +
Sbjct: 383 DVSHNTIS*PSSFPLFTACESTADALD------------FSDSATTISHP--VLSAHISS 246
Query: 106 ASLAS 110
SLA+
Sbjct: 245 *SLAA 231
>TC76719 similar to GP|21595178|gb|AAM66078.1 endo-xyloglucan transferase
putative {Arabidopsis thaliana}, partial (90%)
Length = 1348
Score = 28.1 bits (61), Expect = 5.9
Identities = 17/45 (37%), Positives = 23/45 (50%), Gaps = 8/45 (17%)
Frame = -2
Query: 324 KGTKATAASVGFAP--------VTQSARRPGPVINFIANHPFLFL 360
K +K++++ G AP VT A P NFI NHP L+L
Sbjct: 759 KNSKSSSSLSGPAPFSVHI**AVTTPAESPPTSFNFILNHPNLYL 625
>TC90018 weakly similar to PIR|B84607|B84607 hypothetical protein At2g21950
[imported] - Arabidopsis thaliana, partial (55%)
Length = 965
Score = 27.7 bits (60), Expect = 7.7
Identities = 19/45 (42%), Positives = 25/45 (55%), Gaps = 1/45 (2%)
Frame = +1
Query: 15 SSIHPCLSTLLLASWPP-VQKAAHSTSFSPSSDLTPSTISTPSSL 58
SS+ S L L+S PP + S P S+ +PSTI+T SSL
Sbjct: 145 SSLLRSSSPLALSSLPPNTSSTSLSAHAPPLSNSSPSTITTVSSL 279
>AJ501833 PIR|A87356|A87 sugar ABC transporter ATP-binding protein CC0860
[imported] - Caulobacter crescentus, partial (2%)
Length = 186
Score = 27.7 bits (60), Expect = 7.7
Identities = 15/42 (35%), Positives = 23/42 (54%)
Frame = -2
Query: 30 PPVQKAAHSTSFSPSSDLTPSTISTPSSLRGFPLCSPTKDAA 71
P +Q+ ++F P+S + STI T FP SP+ D+A
Sbjct: 164 PVLQELTLQSAFFPNSRRSSSTIKTWLPPLSFPSSSPSDDSA 39
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,908,336
Number of Sequences: 36976
Number of extensions: 169395
Number of successful extensions: 995
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 985
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 990
length of query: 381
length of database: 9,014,727
effective HSP length: 98
effective length of query: 283
effective length of database: 5,391,079
effective search space: 1525675357
effective search space used: 1525675357
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0348.13


