

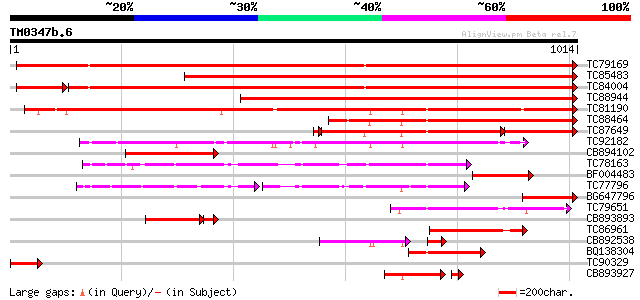
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0347b.6
(1014 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC79169 type IIB calcium ATPase [Medicago truncatula] 1247 0.0
TC85483 type IIB calcium ATPase MCA5 [Medicago truncatula] 1192 0.0
TC84004 type IIB calcium ATPase [Medicago truncatula] 1153 0.0
TC88944 type IIB calcium ATPase [Medicago truncatula] 928 0.0
TC81190 type IIB calcium ATPase [Medicago truncatula] 801 0.0
TC88464 similar to SP|Q9LU41|ACA9_ARATH Potential calcium-transp... 457 e-129
TC87649 similar to SP|Q9SZR1|ACAA_ARATH Potential calcium-transp... 355 e-128
TC92182 type IIA calcium ATPase [Medicago truncatula] 355 6e-98
CB894102 similar to GP|8843813|db Ca2+-transporting ATPase-like ... 190 2e-48
TC78163 H+-ATPase 179 6e-45
BF004483 GP|21314227| type IIB calcium ATPase MCA5 {Medicago tru... 176 4e-44
TC77796 H+-ATPase 121 3e-43
BG647796 GP|21314227|g type IIB calcium ATPase MCA5 {Medicago tr... 156 4e-38
TC79651 similar to PIR|T52581|T52581 Ca2+-transporting ATPase (E... 138 1e-32
CB893893 similar to GP|8843813|db Ca2+-transporting ATPase-like ... 122 2e-31
TC86961 similar to GP|1742951|emb|CAA70946.1 Ca2+-ATPase {Arabid... 129 6e-30
CB892538 similar to SP|Q9XES1|ECA Calcium-transporting ATPase 4 ... 80 1e-25
BQ138304 homologue to GP|6688833|emb putative calcium P-type ATP... 114 2e-25
TC90329 GP|21314227|gb|AAM44081.1 type IIB calcium ATPase MCA5 {... 102 6e-22
CB893927 homologue to GP|17342714|g type IIA calcium ATPase {Med... 85 4e-19
>TC79169 type IIB calcium ATPase [Medicago truncatula]
Length = 3565
Score = 1247 bits (3226), Expect = 0.0
Identities = 635/1005 (63%), Positives = 785/1005 (77%), Gaps = 2/1005 (0%)
Frame = +2
Query: 12 VKSKNSTEEALSKWRKVCGVVKNPKRRFRFTANLSKRYEAAAMRRTNQEKLRVAVLVSKA 71
+K K+ + EALS+WR +VKNP+RRFR A+L+KR A ++ Q K R + V +A
Sbjct: 200 LKDKDRSIEALSRWRSAVSLVKNPRRRFRNVADLAKRALAQEKQKKIQGKFRAVINVQRA 379
Query: 72 AFQFIQGVQPSDYLVPDDVKAAGFHICADELGSIVEGHDVKKLKFHGGVSGIAEKLSTST 131
A F + ++ V + +AAGF I D++ S+V HD K K G V GI KLS S
Sbjct: 380 ALHFTDAIGTPEFKVSEKTRAAGFGIEPDDIASVVRSHDFKNYKKVGEVQGITSKLSVSV 559
Query: 132 TKGLSGDSEARRIRQEVYGINKFAESEVRSFWIFVYEALQDMTLMILAVCAFVSLIVGIA 191
+G+S DS RQE+YG+N++ E +SF +FV++AL D+TL+IL VCA VS+ +G+
Sbjct: 560 DEGVSQDSI--HSRQEIYGLNRYTEKPSKSFLMFVWDALHDLTLIILIVCALVSIGIGLP 733
Query: 192 TEGWPQGSHDGLGIVASILLVVFVTATSDYRQSLQFKDLDKEKKKISIQVTRNGYRQKMS 251
TEGWP+G +DG+GI+ SI LVV VTA SDY+QSLQF DLDKEKKKISI VTR+G RQK+S
Sbjct: 734 TEGWPKGVYDGVGILLSIFLVVTVTAVSDYQQSLQFLDLDKEKKKISIHVTRDGKRQKVS 913
Query: 252 IYNLLPGDLVHLSIGDQVPTDGLFVSGFSVLIDESSLTGESEPVMVTSQNPFLLSGTKVQ 311
IY+L+ GD+VHLS GDQVP DG+F+ G+S+LIDESSL+GESEPV + ++ PFLLSGTKVQ
Sbjct: 914 IYDLVVGDIVHLSTGDQVPADGIFIQGYSLLIDESSLSGESEPVDIDNRRPFLLSGTKVQ 1093
Query: 312 DGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVATLIGKVGLFFAVVTFVVL 371
DG M+VTTVGMRT+WGKLM TLSEGG+DETPLQVKLNGVAT+IGK+GL FAV+TF+VL
Sbjct: 1094 DGQAKMIVTTVGMRTEWGKLMETLSEGGEDETPLQVKLNGVATVIGKIGLTFAVLTFLVL 1273
Query: 372 VKGLMSRKIREGRFWWWSADDAMEMLEFFAIAVTIVVVAVPEGLPLAVTLSLAFAMKKMM 431
+ K G F WS++DA+++L++FAIAVTI+VVA+PEGLPLAVTLSLAFAMKK+M
Sbjct: 1274 TARFVIEKAINGDFTSWSSEDALKLLDYFAIAVTIIVVAIPEGLPLAVTLSLAFAMKKLM 1453
Query: 432 NDKALVRHLAACETMGSATTICSDKTGTLTTNHMTVVKTCICMSSKEVNNKEHG--LCSE 489
ND+ALVRHL+ACETMGSA+ IC+DKTGTLTTNHM V K IC + E+ E L SE
Sbjct: 1454 NDRALVRHLSACETMGSASCICTDKTGTLTTNHMVVDKIWICEKTVEMKGDESTDKLKSE 1633
Query: 490 LPDSAQKLLLQSIFNNTGGEVVVNKRGKREILGTPTESAILEFGLSLGGDPQKERQACKL 549
+ D +LLQ+IF NT EVV + GK+ ILGTPTESA+LEFGL GGD +R++CK+
Sbjct: 1634 ISDEVLSILLQAIFQNTSSEVVKDNEGKQTILGTPTESALLEFGLVSGGDFDAQRRSCKV 1813
Query: 550 VKVEPFNSQKKRMGVVVELPEGGLRAHCKGASEIVLAACDNVIDSKGDVVPLNAESRNYL 609
+KVEPFNS +K+M V+V LP+GG+RA CKGASEIVL CD +IDS G + L E +
Sbjct: 1814 LKVEPFNSDRKKMSVLVGLPDGGVRAFCKGASEIVLKMCDKIIDSNGTTIDLPEEKARIV 1993
Query: 610 ESTIDQFAGEALRTLCLAYIELEHGFSAEDPIPASGYTCIGVVGIKDPVRPGVKESVQVC 669
ID FA EALRTLCLA +++ E IP +GYT I +VGIKDPVRPGVKE+VQ C
Sbjct: 1994 SDIIDGFANEALRTLCLAVKDIDET-QGETNIPENGYTLITIVGIKDPVRPGVKEAVQKC 2170
Query: 670 RSAGIMVRMVTGDNINTAKAIARECGILTEDGLAIEGPDFREKTQEEMFELIPKIQVMAR 729
+AGI VRMVTGDNINTAKAIA+ECGILTE G+AIEGP+FR ++E+M ++IP+IQVMAR
Sbjct: 2171 LAAGISVRMVTGDNINTAKAIAKECGILTEGGVAIEGPEFRNLSEEQMKDIIPRIQVMAR 2350
Query: 730 SSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIIL 789
S PLDKHTLV +LR FGEVVAVTGDGTNDAPALHE+DIGLAMGIAGTEVAKE+ADVII+
Sbjct: 2351 SLPLDKHTLVTRLRNMFGEVVAVTGDGTNDAPALHESDIGLAMGIAGTEVAKENADVIIM 2530
Query: 790 DDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVALLVNFSSAVLTGSAPLTAVQLLWVN 849
DDNF+TIV VAKWGR++YINIQKFVQFQLTVNVVAL+ NF SA +TG+APLTAVQLLWVN
Sbjct: 2531 DDNFTTIVKVAKWGRAIYINIQKFVQFQLTVNVVALITNFVSACITGAAPLTAVQLLWVN 2710
Query: 850 MIMDTLGALALATEPPTDDLMKRAPLGRKGDFINSIMWRNILGQALYQFVVIWFLQTVGK 909
+IMDTLGALALATEPP D LM+R P+GRK FI MWRNI GQ+LYQ +V+ L GK
Sbjct: 2711 LIMDTLGALALATEPPNDGLMERQPVGRKASFITKPMWRNIFGQSLYQLIVLGVLNFEGK 2890
Query: 910 WVFFLRGPNAGVVLNTLIFNSFVFCQVFNEINSREMEEVDVFKGIWDNHVFVAVIGCTVV 969
+ L GP++ VLNTLIFNSFVFCQVFNEINSRE+E++++F+G++D+ +F++VI T V
Sbjct: 2891 RLLGLSGPDSTAVLNTLIFNSFVFCQVFNEINSREIEKINIFRGMFDSWIFLSVILATAV 3070
Query: 970 FQIIIVEYLGTFANTTPLSLVQWIFCLSVGYVGMPIATYLKQIPV 1014
FQ+IIVE+LGTFA+T PL+ W+ L G + MP+A LK IPV
Sbjct: 3071 FQVIIVEFLGTFASTVPLTWQFWLLSLLFGVLSMPLAAILKCIPV 3205
>TC85483 type IIB calcium ATPase MCA5 [Medicago truncatula]
Length = 2541
Score = 1192 bits (3085), Expect = 0.0
Identities = 595/702 (84%), Positives = 653/702 (92%)
Frame = +2
Query: 313 GSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVATLIGKVGLFFAVVTFVVLV 372
GSC MLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVAT+IGK+GLFFA+VTF VLV
Sbjct: 2 GSCKMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVATIIGKIGLFFAIVTFAVLV 181
Query: 373 KGLMSRKIREGRFWWWSADDAMEMLEFFAIAVTIVVVAVPEGLPLAVTLSLAFAMKKMMN 432
+GL+S K+++ FW W+ DDA+EMLE+FAIAVTIVVVAVPEGLPLAVTLSLAFAMKKMMN
Sbjct: 182 QGLVSLKLQQENFWNWNGDDALEMLEYFAIAVTIVVVAVPEGLPLAVTLSLAFAMKKMMN 361
Query: 433 DKALVRHLAACETMGSATTICSDKTGTLTTNHMTVVKTCICMSSKEVNNKEHGLCSELPD 492
DKALVR+LAACETMGSATTICSDKTGTLTTNHMTVVKTCICM SKEV+NK LCSELP+
Sbjct: 362 DKALVRNLAACETMGSATTICSDKTGTLTTNHMTVVKTCICMKSKEVSNKTSSLCSELPE 541
Query: 493 SAQKLLLQSIFNNTGGEVVVNKRGKREILGTPTESAILEFGLSLGGDPQKERQACKLVKV 552
S KLL QSIFNNTGGEVVVNK+GK EILGTPTE+AILEFGLSLGGD Q ERQACKLVKV
Sbjct: 542 SVVKLLQQSIFNNTGGEVVVNKQGKHEILGTPTETAILEFGLSLGGDFQGERQACKLVKV 721
Query: 553 EPFNSQKKRMGVVVELPEGGLRAHCKGASEIVLAACDNVIDSKGDVVPLNAESRNYLEST 612
EPFNS KKRMG VVELP GGLRAHCKGASEIVLAACD V++S G+VVPL+ ES N+L +T
Sbjct: 722 EPFNSTKKRMGAVVELPSGGLRAHCKGASEIVLAACDKVLNSNGEVVPLDEESTNHLTNT 901
Query: 613 IDQFAGEALRTLCLAYIELEHGFSAEDPIPASGYTCIGVVGIKDPVRPGVKESVQVCRSA 672
I+QFA EALRTLCLAY+ELE+GFSAED IP +GYTCIGVVGIKDPVRPGVKESV +CRSA
Sbjct: 902 INQFANEALRTLCLAYMELENGFSAEDTIPVTGYTCIGVVGIKDPVRPGVKESVALCRSA 1081
Query: 673 GIMVRMVTGDNINTAKAIARECGILTEDGLAIEGPDFREKTQEEMFELIPKIQVMARSSP 732
GI VRMVTGDNINTAKAIARECGILT+DG+AIEGP+FREK+ EE+ ELIPKIQVMARSSP
Sbjct: 1082 GITVRMVTGDNINTAKAIARECGILTDDGIAIEGPEFREKSLEELLELIPKIQVMARSSP 1261
Query: 733 LDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDN 792
LDKHTLV+ LRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDN
Sbjct: 1262 LDKHTLVRHLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDN 1441
Query: 793 FSTIVTVAKWGRSVYINIQKFVQFQLTVNVVALLVNFSSAVLTGSAPLTAVQLLWVNMIM 852
FSTIVTVAKWGRSVYINIQKFVQFQLTVN+VAL+VNF+SA LTG+APLTAVQLLWVNMIM
Sbjct: 1442 FSTIVTVAKWGRSVYINIQKFVQFQLTVNIVALIVNFTSACLTGTAPLTAVQLLWVNMIM 1621
Query: 853 DTLGALALATEPPTDDLMKRAPLGRKGDFINSIMWRNILGQALYQFVVIWFLQTVGKWVF 912
DTLGALALATEPP DDLMKRAP+GRKG+FI+++MWRNILGQ+LYQF+VIWFLQ+ GK +F
Sbjct: 1622 DTLGALALATEPPNDDLMKRAPVGRKGNFISNVMWRNILGQSLYQFMVIWFLQSKGKTIF 1801
Query: 913 FLRGPNAGVVLNTLIFNSFVFCQVFNEINSREMEEVDVFKGIWDNHVFVAVIGCTVVFQI 972
L GPN+ +VLNTLIFN+FVFCQVFNEINSREME+++VFKGI DN+VFV VI T+ FQI
Sbjct: 1802 SLDGPNSDLVLNTLIFNAFVFCQVFNEINSREMEKINVFKGILDNYVFVGVISATIFFQI 1981
Query: 973 IIVEYLGTFANTTPLSLVQWIFCLSVGYVGMPIATYLKQIPV 1014
IIVEYLGTFANTTPL+LVQW FCL VG++GMPIA LK+IPV
Sbjct: 1982 IIVEYLGTFANTTPLTLVQWFFCLFVGFMGMPIAARLKKIPV 2107
>TC84004 type IIB calcium ATPase [Medicago truncatula]
Length = 3655
Score = 1153 bits (2982), Expect(2) = 0.0
Identities = 581/914 (63%), Positives = 723/914 (78%), Gaps = 4/914 (0%)
Frame = +1
Query: 105 IVEGHDVKKLKFHGGVSGIAEKLSTSTTKGLSGDSEARRIRQEVYGINKFAESEVRSFWI 164
+ D K L +GGV +A KLS S +G++ S RQ+++G N++ E R+F +
Sbjct: 547 LFRSQDYKNLSNNGGVEAVARKLSVSIDEGVNDTSVD--CRQQIFGANRYTEKPSRTFLM 720
Query: 165 FVYEALQDMTLMILAVCAFVSLIVGIATEGWPQGSHDGLGIVASILLVVFVTATSDYRQS 224
FV++ALQD+TL IL VCA VS+ +G+ATEGWP+G++DG+GI+ SI LVV VTA SDYRQS
Sbjct: 721 FVWDALQDLTLTILMVCAVVSIGIGLATEGWPKGTYDGVGIILSIFLVVIVTAVSDYRQS 900
Query: 225 LQFKDLDKEKKKISIQVTRNGYRQKMSIYNLLPGDLVHLSIGDQVPTDGLFVSGFSVLID 284
LQF DLD+EKKKI +QV R+G R+K+SIY+++ GD++HLS GDQVP DG+++SG+S+LID
Sbjct: 901 LQFMDLDREKKKIFVQVNRDGKRKKISIYDVVVGDIIHLSTGDQVPADGIYISGYSLLID 1080
Query: 285 ESSLTGESEPVMVTSQNPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETP 344
ESSL+GESEPV +T ++PFLLSGTKVQDG MLVTTVGMRT+WGKLM TL+EGG+DETP
Sbjct: 1081 ESSLSGESEPVFITEEHPFLLSGTKVQDGQGKMLVTTVGMRTEWGKLMETLNEGGEDETP 1260
Query: 345 LQVKLNGVATLIGKVGLFFAVVTFVVLVKGLMSRKIREGRFWWWSADDAMEMLEFFAIAV 404
LQVKLNGVAT+IGK+GLFFA+VTF+VL + K G F WS++DA ++L+FFAIAV
Sbjct: 1261 LQVKLNGVATIIGKIGLFFAIVTFLVLTVRFLVEKALHGEFGNWSSNDATKLLDFFAIAV 1440
Query: 405 TIVVVAVPEGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNH 464
TI+VVAVPEGLPLAVTLSLAFAMKK+MND ALVRHL+ACETMGSA+ IC+DKTGTLTTNH
Sbjct: 1441 TIIVVAVPEGLPLAVTLSLAFAMKKLMNDMALVRHLSACETMGSASCICTDKTGTLTTNH 1620
Query: 465 MTVVKTCICMSSKEVNNKEHG--LCSELPDSAQKLLLQSIFNNTGGEVVVNKRGKREILG 522
M V K IC ++ ++ E L + + + +LLQ+IF NT EVV +K GK ILG
Sbjct: 1621 MVVNKIWICENTTQLKGDESADELKTNISEGVLSILLQAIFQNTSAEVVKDKNGKNTILG 1800
Query: 523 TPTESAILEFGLSLGG--DPQKERQACKLVKVEPFNSQKKRMGVVVELPEGGLRAHCKGA 580
+PTESA+LEFGL LG D + +A K++K+EPFNS +K+M V+V LP G ++A CKGA
Sbjct: 1801 SPTESALLEFGLLLGSEFDARNHSKAYKILKLEPFNSVRKKMSVLVGLPNGRVQAFCKGA 1980
Query: 581 SEIVLAACDNVIDSKGDVVPLNAESRNYLESTIDQFAGEALRTLCLAYIELEHGFSAEDP 640
SEI+L CD +ID G+VV L A+ N + I+ FA EALRTLCLA ++ E
Sbjct: 1981 SEIILEMCDKMIDCNGEVVDLPADRANIVSDVINSFASEALRTLCLAVRDINET-QGETN 2157
Query: 641 IPASGYTCIGVVGIKDPVRPGVKESVQVCRSAGIMVRMVTGDNINTAKAIARECGILTED 700
IP SGYT I +VGIKDPVRPGVKE+VQ C +AGI VRMVTGDNINTAKAIA+ECGILT+D
Sbjct: 2158 IPDSGYTLIALVGIKDPVRPGVKEAVQTCIAAGITVRMVTGDNINTAKAIAKECGILTDD 2337
Query: 701 GLAIEGPDFREKTQEEMFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDA 760
G+AIEGP FRE + E+M ++IP+IQVMARS PLDKH LV LR FGEVVAVTGDGTNDA
Sbjct: 2338 GVAIEGPSFRELSDEQMKDIIPRIQVMARSLPLDKHKLVTNLRNMFGEVVAVTGDGTNDA 2517
Query: 761 PALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTV 820
PALHEADIGLAMGIAGTEVAKE ADVII+DDNF+TIV V KWGR+VYINIQKFVQFQLTV
Sbjct: 2518 PALHEADIGLAMGIAGTEVAKEKADVIIMDDNFATIVNVVKWGRAVYINIQKFVQFQLTV 2697
Query: 821 NVVALLVNFSSAVLTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKRAPLGRKGD 880
NVVAL++NF SA +TGSAPLTAVQLLWVN+IMDTLGALALATEPP D L+KR P+GR
Sbjct: 2698 NVVALIINFVSACITGSAPLTAVQLLWVNLIMDTLGALALATEPPNDGLLKRPPVGRGAS 2877
Query: 881 FINSIMWRNILGQALYQFVVIWFLQTVGKWVFFLRGPNAGVVLNTLIFNSFVFCQVFNEI 940
FI MWRNI+GQ++YQ +V+ L GK + + G +A VLNTLIFNSFVFCQVFNEI
Sbjct: 2878 FITKTMWRNIIGQSIYQLIVLAILNFDGKRLLGINGSDATEVLNTLIFNSFVFCQVFNEI 3057
Query: 941 NSREMEEVDVFKGIWDNHVFVAVIGCTVVFQIIIVEYLGTFANTTPLSLVQWIFCLSVGY 1000
NSR++E++++F+G++D+ +F+ +I TV FQ++IVE+LG FA+T PLS W+ + +G
Sbjct: 3058 NSRDIEKINIFRGMFDSWIFLLIIFSTVAFQVVIVEFLGAFASTVPLSWQLWLLSVLIGA 3237
Query: 1001 VGMPIATYLKQIPV 1014
+ MP+A +K IPV
Sbjct: 3238 ISMPLAVIVKCIPV 3279
Score = 71.2 bits (173), Expect(2) = 0.0
Identities = 37/91 (40%), Positives = 56/91 (60%)
Frame = +2
Query: 12 VKSKNSTEEALSKWRKVCGVVKNPKRRFRFTANLSKRYEAAAMRRTNQEKLRVAVLVSKA 71
++ KN + EAL +WR +VKN +RRFR A+L KR EA +++ +EK+R+A+ V KA
Sbjct: 266 LEPKNRSVEALRRWRSAVTLVKNRRRRFRMVADLEKRSEAEQIKQGIKEKIRIALYVQKA 445
Query: 72 AFQFIQGVQPSDYLVPDDVKAAGFHICADEL 102
A QFI +Y + + AGF I +E+
Sbjct: 446 ALQFIDAGNRVEYKLSREAIEAGFDIHPNEI 538
>TC88944 type IIB calcium ATPase [Medicago truncatula]
Length = 1883
Score = 928 bits (2399), Expect = 0.0
Identities = 464/604 (76%), Positives = 531/604 (87%), Gaps = 2/604 (0%)
Frame = +3
Query: 413 EGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNHMTVVKTCI 472
+GLPLAVTL LAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNHMTVVK CI
Sbjct: 3 DGLPLAVTLILAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNHMTVVKACI 182
Query: 473 CMSSKEVNNK--EHGLCSELPDSAQKLLLQSIFNNTGGEVVVNKRGKREILGTPTESAIL 530
C KEVN+ S+LPDSA +LL+SIFNNTGGEVV N+ GK EILG+PTE+AIL
Sbjct: 183 CGKIKEVNSSIDSSDFSSDLPDSAIAILLESIFNNTGGEVVKNENGKIEILGSPTETAIL 362
Query: 531 EFGLSLGGDPQKERQACKLVKVEPFNSQKKRMGVVVELPEGGLRAHCKGASEIVLAACDN 590
EFGLSLGGD KERQA KLVKVEPFNS KKRMGVV++LP+GG RAHCKGASEI+LAACD
Sbjct: 363 EFGLSLGGDFHKERQASKLVKVEPFNSIKKRMGVVLQLPDGGYRAHCKGASEIILAACDK 542
Query: 591 VIDSKGDVVPLNAESRNYLESTIDQFAGEALRTLCLAYIELEHGFSAEDPIPASGYTCIG 650
+DS +VPL+ +S ++L TI++FA EALRTLCLAYI++ F PIP +GYTC+G
Sbjct: 543 FVDSNSKIVPLDEDSISHLNDTIEKFANEALRTLCLAYIDIHDEFLVGSPIPVNGYTCVG 722
Query: 651 VVGIKDPVRPGVKESVQVCRSAGIMVRMVTGDNINTAKAIARECGILTEDGLAIEGPDFR 710
+VGIKDPVRPGV+ESV +CRSAGI VRMVTGDNINTAKAIARECGILT DG+AIEGP+FR
Sbjct: 723 IVGIKDPVRPGVRESVAICRSAGITVRMVTGDNINTAKAIARECGILT-DGIAIEGPEFR 899
Query: 711 EKTQEEMFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGL 770
E +++E+ ++IPKIQVMARSSP+DKHTLVK LRTTF EVVAVTGDGTNDAPALHEADIGL
Sbjct: 900 EMSEKELLDIIPKIQVMARSSPMDKHTLVKHLRTTFEEVVAVTGDGTNDAPALHEADIGL 1079
Query: 771 AMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVALLVNFS 830
AMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVAL+VNF+
Sbjct: 1080 AMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVALIVNFT 1259
Query: 831 SAVLTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKRAPLGRKGDFINSIMWRNI 890
SA LTG+APLTAVQLLWVNMIMDTLGALALATEPP D+LMKRAP+GRKG+FI+++MWRNI
Sbjct: 1260 SACLTGNAPLTAVQLLWVNMIMDTLGALALATEPPNDELMKRAPVGRKGNFISNVMWRNI 1439
Query: 891 LGQALYQFVVIWFLQTVGKWVFFLRGPNAGVVLNTLIFNSFVFCQVFNEINSREMEEVDV 950
GQ++YQFV+IW LQT GK VF L GP++ ++LNTLIFNSFVFCQVFNEI+SR+ME ++V
Sbjct: 1440 TGQSIYQFVIIWLLQTRGKTVFHLDGPDSDLILNTLIFNSFVFCQVFNEISSRDMERINV 1619
Query: 951 FKGIWDNHVFVAVIGCTVVFQIIIVEYLGTFANTTPLSLVQWIFCLSVGYVGMPIATYLK 1010
F+GI N+VF AV+ CT +FQIIIVE+LGT+ANT+PLSL W+ + +G +GMPI LK
Sbjct: 1620 FEGILKNYVFTAVLTCTAIFQIIIVEFLGTYANTSPLSLKLWLISVFLGVLGMPIGAALK 1799
Query: 1011 QIPV 1014
IPV
Sbjct: 1800 MIPV 1811
>TC81190 type IIB calcium ATPase [Medicago truncatula]
Length = 3277
Score = 801 bits (2068), Expect = 0.0
Identities = 466/1029 (45%), Positives = 641/1029 (62%), Gaps = 41/1029 (3%)
Frame = +1
Query: 27 KVCGVVKNPKRRFRF--TANLSKRY-----EAAAMRRTNQEKLRVAVLVSKAAFQFIQGV 79
KV N KRR+RF TA S+R + R+ + ++ S ++ + +
Sbjct: 70 KVTSKYTNAKRRWRFAYTAIYSRRVMLSLAKEVISRKNSNPYTKLFHTESSSSTTTLDII 249
Query: 80 QPSDYLVPDDVKAAGFHICAD------ELGSIVEGHDVKKLKFHGGVSGIAEKLSTSTTK 133
+P L+ + + +D +L +V+ ++K L GGV G+ L T TK
Sbjct: 250 EP---LITQHNGTNHYSLVSDVVVDKTKLADMVKDKNLKSLSEFGGVEGVGHVLGTFPTK 420
Query: 134 GLSGDSEARRIRQEVYGINKFAESEVRSFWIFVYEALQDMTLMILAVCAFVSLIVGIATE 193
G+ G + R E++G N + + + FV EA D T++IL VCA +SL GI
Sbjct: 421 GIIGSDDDISRRLELFGSNTYKKPPPKGLLHFVLEAFNDTTIIILLVCAGLSLGFGIKEH 600
Query: 194 GWPQGSHDGLGIVASILLVVFVTATSDYRQSLQFKDLDKEKKKISIQVTRNGYRQKMSIY 253
G +G ++G I ++ LVV V+A S++RQ QF L K I ++V RNG Q++SI+
Sbjct: 601 GPGEGWYEGGSIFLAVFLVVVVSALSNFRQERQFHKLSKISNNIKVEVVRNGRPQQISIF 780
Query: 254 NLLPGDLVHLSIGDQVPTDGLFVSGFSVLIDESSLTGESEPVMVTS-QNPFLLSGTKVQD 312
++L GD+V L IGDQ+P DG+F+SG+S+ +DESS+TGES+ V + + PFLLSG KV D
Sbjct: 781 DVLVGDIVSLKIGDQIPADGVFLSGYSLQVDESSMTGESDHVEIEPLRAPFLLSGAKVVD 960
Query: 313 GSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVATLIGKVGLFFAVVTFVVLV 372
G MLVT+VG T WG++M+++S ++ TPLQ +L+ + + IGKVGL A + +VL+
Sbjct: 961 GYAQMLVTSVGKNTSWGQMMSSISRDTNERTPLQARLDKLTSSIGKVGLAVAFLVLLVLL 1140
Query: 373 KGLMS------RKIREGRFWWWSADDAME-MLEFFAIAVTIVVVAVPEGLPLAVTLSLAF 425
+ + +E R +D M ++ A AVTIVVVA+PEGLPLAVTL+LA+
Sbjct: 1141 IRYFTGNSHDEKGNKEFRGSKTDINDVMNSVVSIVAAAVTIVVVAIPEGLPLAVTLTLAY 1320
Query: 426 AMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNHMTVVKTCICMSSKEVNNKEHG 485
+MK+MM D A+VR L+ACETMGSAT IC+DKTGTLT N M V K C+ N
Sbjct: 1321 SMKRMMADHAMVRKLSACETMGSATVICTDKTGTLTLNQMRVTKFCL-----GPENIIEN 1485
Query: 486 LCSELPDSAQKLLLQSIFNNTGGEVVVNKRGKR-EILGTPTESAILEFG-LSLGGDPQKE 543
+ + +L Q + NT G V G EI G+PTE AIL + L LG D +
Sbjct: 1486 FSNAMTPKVLELFHQGVGLNTTGSVYNPPSGSEPEISGSPTEKAILMWAVLDLGMDMDEM 1665
Query: 544 RQACKLVKVEPFNSQKKRMGVVV--ELPEGGLRAHCKGASEIVLAACDNVIDSKGDVVPL 601
+Q K++ VE FNS+KKR GV + E + + H KGA+E++LA C N IDS G L
Sbjct: 1666 KQKHKVLHVETFNSEKKRSGVAIRKENDDNSVHVHWKGAAEMILAMCTNYIDSNGARKSL 1845
Query: 602 NAESRNYLESTIDQFAGEALRTLCLAYIELEHGFSAEDPIPAS----------GYTCIGV 651
+ E R+ +E I A +LR + A+ E+ + I G T +G+
Sbjct: 1846 DEEERSKIERIIQVMAASSLRCIAFAHTEISDSEDIDYMIKREKKSHQMLREDGLTLLGI 2025
Query: 652 VGIKDPVRPGVKESVQVCRSAGIMVRMVTGDNINTAKAIARECGILTED------GLAIE 705
VG+KDP RP K++V+ C++AG+ ++M+TGDNI TAKAIA ECGIL + G +E
Sbjct: 2026 VGLKDPCRPNTKKAVETCKAAGVEIKMITGDNIFTAKAIAIECGILDSNSDHAKAGEVVE 2205
Query: 706 GPDFREKTQEEMFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHE 765
G +FR T+EE E + I+VMARSSP+DK +V+ LR G VVAVTGDGTNDAPAL E
Sbjct: 2206 GVEFRSYTEEERMEKVDNIRVMARSSPMDKLLMVQCLRKK-GHVVAVTGDGTNDAPALKE 2382
Query: 766 ADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVAL 825
ADIGL+MGI GTEVAKES+D++ILDDNF+++ TV +WGR VY NIQKF+QFQLTVNV AL
Sbjct: 2383 ADIGLSMGIQGTEVAKESSDIVILDDNFNSVATVLRWGRCVYNNIQKFIQFQLTVNVAAL 2562
Query: 826 LVNFSSAVLTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKRAPLGRKGDFINSI 885
++NF +AV +G PLT VQLLWVN+IMDTLGALALATE PT +LMK+ P+GR I +I
Sbjct: 2563 VINFIAAVSSGDVPLTTVQLLWVNLIMDTLGALALATERPTKELMKKKPIGRTAPLITNI 2742
Query: 886 MWRNILGQALYQFVVIWFLQTVGKWVFFLRGPNAGVVLNTLIFNSFVFCQVFNEINSREM 945
MWRN+L QA YQ V+ +Q GK +F + + V +TLIFN+FV CQVFNE NSR M
Sbjct: 2743 MWRNLLAQASYQIAVLLIMQFYGKSIFNV----SKEVKDTLIFNTFVLCQVFNEFNSRSM 2910
Query: 946 EEVDVFKGIWDNHVFVAVIGCTVVFQIIIVEYLGTFANTTPLSLVQWIFCLSVGYVGMPI 1005
E++ VF+GI NH+F+ +IG T+V QI++VE L FA+T L+ QW C+ + V P+
Sbjct: 2911 EKLYVFEGILKNHLFLGIIGITIVLQILMVELLRKFADTERLTWEQWGICIGIAVVSWPL 3090
Query: 1006 ATYLKQIPV 1014
A +K IPV
Sbjct: 3091 ACLVKLIPV 3117
>TC88464 similar to SP|Q9LU41|ACA9_ARATH Potential calcium-transporting ATPase
9 plasma membrane-type (EC 3.6.3.8) (Ca2+-ATPase
isoform 9)., partial (43%)
Length = 1608
Score = 457 bits (1176), Expect = e-129
Identities = 245/459 (53%), Positives = 319/459 (69%), Gaps = 14/459 (3%)
Frame = +3
Query: 570 EGGLRAHCKGASEIVLAACDNVIDSKGDVVPLNAESRNYLESTIDQFAGEALRTLCLAY- 628
+ G+ H KGA+EIVL AC +DS + + E +++L+ ID A +LR + +AY
Sbjct: 6 DSGVHIHWKGAAEIVLGACTQYLDSNSHLQSIEQE-KDFLKEAIDDMAARSLRCVAIAYR 182
Query: 629 -IELEHGFSAEDPI-----PASGYTCIGVVGIKDPVRPGVKESVQVCRSAGIMVRMVTGD 682
EL+ S E+ + P + +VGIKDP RPGVK++V++C AG+ VRMVTGD
Sbjct: 183 SYELDKIPSNEEDLAQWSLPEDELVLLAIVGIKDPCRPGVKDAVRICTDAGVKVRMVTGD 362
Query: 683 NINTAKAIARECGILTE-----DGLAIEGPDFREKTQEEMFELIPKIQVMARSSPLDKHT 737
N+ TAKAIA ECGIL D IEG FRE +++E ++ KI VM RSSP DK
Sbjct: 363 NLQTAKAIALECGILASNEEAVDPCIIEGKVFRELSEKEREQVAKKITVMGRSSPNDKLL 542
Query: 738 LVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIV 797
LV+ LR G+VVAVTGDGTNDAPALHEADIGL+MGI GTEVAKES+D+IILDDNF+++V
Sbjct: 543 LVQALRKG-GDVVAVTGDGTNDAPALHEADIGLSMGIQGTEVAKESSDIIILDDNFASVV 719
Query: 798 TVAKWGRSVYINIQKFVQFQLTVNVVALLVNFSSAVLTGSAPLTAVQLLWVNMIMDTLGA 857
V +WGRSVY NIQKF+QFQLTVNV AL++N +A+ +G PL AVQLLWVN+IMDTLGA
Sbjct: 720 KVVRWGRSVYANIQKFIQFQLTVNVAALVINVVAAISSGDVPLNAVQLLWVNLIMDTLGA 899
Query: 858 LALATEPPTDDLMKRAPLGRKGDFINSIMWRNILGQALYQFVVIWFLQTVGKWVFFLRGP 917
LALATEPPTD LM R+P+GR+ I +IMWRN++ QALYQ V+ FL G+ + +
Sbjct: 900 LALATEPPTDHLMHRSPVGRREPLITNIMWRNLIVQALYQVSVLLFLNFCGESILPKQDT 1079
Query: 918 NAG--VVLNTLIFNSFVFCQVFNEINSREMEEVDVFKGIWDNHVFVAVIGCTVVFQIIIV 975
A V NTLIFN+FV CQ+FNE N+R+ +E++VF G+ N +F+ +IG T + QI+I+
Sbjct: 1080 KAHDYQVKNTLIFNAFVMCQIFNEFNARKPDEMNVFPGVTKNRLFMGIIGITFILQIVII 1259
Query: 976 EYLGTFANTTPLSLVQWIFCLSVGYVGMPIATYLKQIPV 1014
E+LG FA+T L W+ L +G V P+A K IPV
Sbjct: 1260 EFLGKFASTVRLDWKLWLVSLIIGLVSWPLAIAGKFIPV 1376
>TC87649 similar to SP|Q9SZR1|ACAA_ARATH Potential calcium-transporting
ATPase 10 plasma membrane-type (EC 3.6.3.8), partial
(43%)
Length = 2159
Score = 355 bits (910), Expect(3) = e-128
Identities = 188/341 (55%), Positives = 236/341 (69%), Gaps = 12/341 (3%)
Frame = +3
Query: 558 QKKRMGVVVELPEGGLRAHCKGASEIVLAACDNVIDSKGDVVPLNAESRNYLESTIDQFA 617
+K+ GV ++ + + H KGA+EIVLA C ID+ +V ++ E + + I+ A
Sbjct: 60 RKREAGVAIQTADSDVHIHWKGAAEIVLACCTGYIDANDQLVEIDEEKMTFFKKAIEDMA 239
Query: 618 GEALRTLCLAYIELE-------HGFSAEDPIPASGYTCIGVVGIKDPVRPGVKESVQVCR 670
++LR + +AY E A+ +P + +VGIKDP RPGVK SVQ+C+
Sbjct: 240 SDSLRCVAIAYRPYEKEKVPDNEEQLADWSLPEEELVLLAIVGIKDPCRPGVKNSVQLCQ 419
Query: 671 SAGIMVRMVTGDNINTAKAIARECGILTE-----DGLAIEGPDFREKTQEEMFELIPKIQ 725
AG+ V+MVTGDN+ TAKAIA ECGIL+ + IEG FR + E E+ I
Sbjct: 420 KAGVKVKMVTGDNVKTAKAIALECGILSSLADVTERSVIEGKTFRALSDSEREEIAESIS 599
Query: 726 VMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESAD 785
VM RSSP DK LV+ LR G VVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKES+D
Sbjct: 600 VMGRSSPNDKLLLVQALRRK-GHVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESSD 776
Query: 786 VIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVALLVNFSSAVLTGSAPLTAVQL 845
+IILDDNF+++V V +WGRSVY NIQKF+QFQLTVNV AL++N +AV +G PL AVQL
Sbjct: 777 IIILDDNFASVVKVVRWGRSVYANIQKFIQFQLTVNVAALVINVVAAVSSGDVPLNAVQL 956
Query: 846 LWVNMIMDTLGALALATEPPTDDLMKRAPLGRKGDFINSIM 886
LWVN+IMDTLGALALATEPPTD LM R+P+GR+ I +IM
Sbjct: 957 LWVNLIMDTLGALALATEPPTDHLMDRSPVGRREPLITNIM 1079
Score = 121 bits (304), Expect(3) = e-128
Identities = 60/133 (45%), Positives = 89/133 (66%), Gaps = 3/133 (2%)
Frame = +1
Query: 885 IMWRNILGQALYQFVVIWFLQTVGKWVFFLRGP---NAGVVLNTLIFNSFVFCQVFNEIN 941
+ WRN+L QA+YQ V+ L G + L +A V NTLIFN+FV CQ+FNE N
Sbjct: 1075 LWWRNLLIQAMYQVSVLLVLNFRGISILGLEHQPTEHAIKVKNTLIFNAFVICQIFNEFN 1254
Query: 942 SREMEEVDVFKGIWDNHVFVAVIGCTVVFQIIIVEYLGTFANTTPLSLVQWIFCLSVGYV 1001
+R+ +E ++FKG+ N++F+ ++G TVV Q+IIVE+LG F TT L+ QW+ +++G++
Sbjct: 1255 ARKPDEYNIFKGVTRNYLFMGIVGFTVVLQVIIVEFLGKFTTTTRLNWKQWLISVAIGFI 1434
Query: 1002 GMPIATYLKQIPV 1014
G P+A K IPV
Sbjct: 1435 GWPLAVVGKLIPV 1473
Score = 22.3 bits (46), Expect(3) = e-128
Identities = 9/18 (50%), Positives = 12/18 (66%)
Frame = +2
Query: 544 RQACKLVKVEPFNSQKKR 561
R ++ V PFNS+KKR
Sbjct: 17 RSESSILHVFPFNSEKKR 70
>TC92182 type IIA calcium ATPase [Medicago truncatula]
Length = 3148
Score = 355 bits (910), Expect = 6e-98
Identities = 283/895 (31%), Positives = 444/895 (48%), Gaps = 91/895 (10%)
Frame = +1
Query: 125 EKLSTSTTKGLSGDSEARRIRQEVYGINKFAESEVRSFWIFVYEALQDMTLMILAVCAFV 184
E+ KGLS + +R +E YG N+ A+ + + W V E DM + IL AF+
Sbjct: 52 EEYGVKLEKGLSSNEVQKR--REKYGWNELAKEKGKPLWKLVLEQFDDMLVKILLAAAFI 225
Query: 185 SLIVGIATEGWPQGSHDGLGIVASILLVVFVTATSDYRQSLQFKDLD--KEKKKISIQVT 242
S ++ EG G + + IL++V ++++ K L+ KE + SI+V
Sbjct: 226 SFLLAYF-EGSESGFEAYVEPLVIILILVLNAIVGVWQENNAEKALEALKELQCESIKVL 402
Query: 243 RNGYR-QKMSIYNLLPGDLVHLSIGDQVPTDGLFVS--GFSVLIDESSLTGESEPVM--- 296
R+GY + L+PGD+V L +GD+VP D + ++ +++SSLTGE+ PV+
Sbjct: 403 RDGYFVPDLPARELVPGDIVELRVGDKVPADMRVAALKTSTLRLEQSSLTGEAMPVLKGT 582
Query: 297 ---------VTSQNPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGG--DDETPL 345
+ ++ + +GT V +GSC +V T M T+ GK+ + E + +TPL
Sbjct: 583 NPIFMDDCELQAKENMVFAGTTVVNGSCICIVITTAMNTEIGKIQKQIHEASLEESDTPL 762
Query: 346 QVKLNGVA-TLIGKVGLFFAVVTFVVLVKGLMSRKIREGRFWWWSADDAMEMLE---FFA 401
+ KL+ L +G+ VV +++ K +S + +G W + + +F
Sbjct: 763 KKKLDEFGGRLTTSIGIVCLVV-WIINYKNFISWDVVDG----WPTNIQFSFQKCTYYFK 927
Query: 402 IAVTIVVVAVPEGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLT 461
IAV + V A+PEGLP +T LA +KM A+VR L + ET+G T ICSDKTGTLT
Sbjct: 928 IAVALAVAAIPEGLPAVITTCLALGTRKMAQKNAIVRKLPSVETLGCTTVICSDKTGTLT 1107
Query: 462 TNHMTVV-------KTCIC----MSSKEVNNKEHGLCSELPDSAQKLLLQ-----SIFNN 505
TN M+ KT C + + K+ G+ + LL ++ N+
Sbjct: 1108TNQMSATEFFTLGGKTTACRVISVEGTTYDPKDGGIVDWTCYNMDANLLAMAEICAVCND 1287
Query: 506 TGGEVVVNKRGKREILGTPTESAILEFGLSLGGDPQKER--------------------- 544
G V + R R G PTE+A+ +G K R
Sbjct: 1288AG--VYFDGRLFRAT-GLPTEAALKVLVEKMGFPDTKSRNKTHDALVATNNMVDCNTLKL 1458
Query: 545 -------QACKLVKVEPFNSQKKRMGVVVELPEGGLRAHCKGASEIVLAACDNVIDSKGD 597
+ K V F+ +K M V+V P+G R KGA E +L V + G
Sbjct: 1459GCCEWWNRRSKRVATLEFDRVRKSMSVIVREPDGQNRLLVKGAVESLLERSSYVQLADGS 1638
Query: 598 VVPLNAESRNYLESTIDQFAGEALRTLCLAYIELEHGFSA--EDPIPA------------ 643
+VP++ + R L + + + + LR L LA + FS D PA
Sbjct: 1639LVPIDDQCRELLLQRLHEMSSKGLRCLGLACKDELGEFSDYYADTHPAHKKLLDPTYYSS 1818
Query: 644 --SGYTCIGVVGIKDPVRPGVKESVQVCRSAGIMVRMVTGDNINTAKAIARECGILTED- 700
S +GVVG++DP R V ++++ C+ AGI V ++TGDN +TA+AI +E + + D
Sbjct: 1819IESDLIFVGVVGLRDPPREEVHKAIEDCKQAGIRVMVITGDNKSTAEAICKEIKLFSTDE 1998
Query: 701 ---GLAIEGPDFREKTQEEMFELIPKI--QVMARSSPLDKHTLVKQLRTTFGEVVAVTGD 755
G ++ G +F + E +L+ + +V +R+ P K +V+ L+ GE+VA+TGD
Sbjct: 1999DLTGQSLTGKEFMSLSHSEQVKLLLRNGGKVFSRAEPRHKQEIVRLLKE-MGEIVAMTGD 2175
Query: 756 GTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFVQ 815
G NDAPAL ADIG+AMGI GTEVAKE++D+++ DDNFSTIV+ GR++Y N++ F++
Sbjct: 2176GVNDAPALKLADIGIAMGITGTEVAKEASDMVLADDNFSTIVSAIAEGRAIYNNMKAFIR 2355
Query: 816 FQLTVNVVALLVNFSSAVLTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKRAPL 875
+ ++ NV ++ F +A L + VQLLWVN++ D A AL P D+M++ P
Sbjct: 2356YMISSNVGEVISIFLTAALGIPECMIPVQLLWVNLVTDGPPATALGFNPADVDIMQKPP- 2532
Query: 876 GRKGDFINSIMWRNILGQALYQFVVI-WFL-QTVGKWVFFLRGPNAGVVLNTLIF 928
RK D W L++++V WFL + W F +V++T IF
Sbjct: 2533-RKSDDALISAW------VLFRYLVSNWFLCRNCNSWHFC-------IVVHTSIF 2655
>CB894102 similar to GP|8843813|db Ca2+-transporting ATPase-like protein
{Arabidopsis thaliana}, partial (15%)
Length = 802
Score = 190 bits (483), Expect = 2e-48
Identities = 94/165 (56%), Positives = 132/165 (79%)
Frame = +2
Query: 208 SILLVVFVTATSDYRQSLQFKDLDKEKKKISIQVTRNGYRQKMSIYNLLPGDLVHLSIGD 267
+++LV+ VTA SDY+QSLQF+DL++EK+ I ++V R G R ++SIY+L+ GD++ L+IG+
Sbjct: 5 AVILVIVVTAVSDYKQSLQFRDLNEEKRNIHLEVIRGGRRVEISIYDLVVGDVIPLNIGN 184
Query: 268 QVPTDGLFVSGFSVLIDESSLTGESEPVMVTSQNPFLLSGTKVQDGSCTMLVTTVGMRTQ 327
QVP DG+ ++G S+ IDESS+TGES+ V S++PF++SG KV DGS TMLVT VG+ T+
Sbjct: 185 QVPADGVVITGHSLSIDESSMTGESKIVHKDSKDPFMMSGCKVADGSGTMLVTGVGINTE 364
Query: 328 WGKLMATLSEGGDDETPLQVKLNGVATLIGKVGLFFAVVTFVVLV 372
WG LMA++SE +ETPLQV+LNGVAT IG VGL AV+ +VL+
Sbjct: 365 WGLLMASISEDTGEETPLQVRLNGVATFIGIVGLSVAVLVLIVLL 499
>TC78163 H+-ATPase
Length = 3295
Score = 179 bits (453), Expect = 6e-45
Identities = 177/710 (24%), Positives = 310/710 (42%), Gaps = 14/710 (1%)
Frame = +1
Query: 131 TTKGLSGDSEARRIRQEVYGINKFAE---SEVRSFWIFVYEALQDMTLMILAVCAFVSLI 187
T +GL+ + R+ E++G NK E S++ F F++ L + A+ A
Sbjct: 223 TKEGLTCEEVQERL--ELFGYNKLEEKKESKILKFLGFMWNPLS-WVMEAAAIMAIAMAH 393
Query: 188 VGIATEGWP-QGSH-DGLGIVASILLVVFVTAT-------SDYRQSLQFKDLDKEKKKIS 238
G +G QG + D +GI+ ILL++ T + + +L + K K
Sbjct: 394 GGRNMDGTKKQGDYQDFVGII--ILLIINSTISFIEENNAGNAAAALMARLAPKAK---- 555
Query: 239 IQVTRNGYRQKMSIYNLLPGDLVHLSIGDQVPTDGLFVSGFSVLIDESSLTGESEPVMVT 298
V R+G + L+PGD+V + +GD +P D + G + ID+S+LTGES PV
Sbjct: 556 --VLRDGKWSEEDASVLVPGDIVSIKLGDIIPADARLLEGDPLKIDQSALTGESLPV-TK 726
Query: 299 SQNPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVATLIGK 358
+ SG+ + G +V G+ T +GK A L E Q L +
Sbjct: 727 HPGEGIYSGSTCKQGEIEAIVIATGVHTFFGKA-AHLVENTTHVGHFQKVLTSIGNFC-I 900
Query: 359 VGLFFAVVTFVVLVKGLMSRKIREGRFWWWSADDAMEMLEFFAIAVTIVVVAVPEGLPLA 418
+ +V ++++ G+ + R G D+ + +L + +P +P
Sbjct: 901 CSIAIGMVIEIIVIYGVHGKGYRNG------IDNLLVLL----------IGGIPIAMPTV 1032
Query: 419 VTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNHMTVVKTCICMSSKE 478
+++++A K+ A+ + + A E M +CSDKTGTLT N +TV K I + +K
Sbjct: 1033LSVTMAIGSHKLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLTVDKDMIEVFAKG 1212
Query: 479 VNNKEHGLCSELPDSAQKLLLQSIFNNTGGEVVVNKRGKREILGTPTESAILEFGLSLGG 538
V+ ++VV + L + AI +S+
Sbjct: 1213VDK---------------------------DLVVLMAARASRL--ENQDAIDCAIVSMLA 1305
Query: 539 DPQKERQACKLVKVEPFNSQKKRMGVVVELPEGGLRAHCKGASEIVLAACDNVIDSKGDV 598
DP++ R K V PFN KR + G + KGA E +L N
Sbjct: 1306DPKEARTGIKEVHFLPFNPTDKRTALTYIDAAGNMHRVSKGAPEQILNLARN-------- 1461
Query: 599 VPLNAESRNYLESTIDQFAGEALRTLCLAYIELEHGFSAEDPIPASGYTCIGVVGIKDPV 658
AE + S ID+FA LR+L +A E+ G P + + ++ + DP
Sbjct: 1462---KAEIAQKVHSMIDKFAERGLRSLGVARQEVPEGSKDS---PGGPWEFVALLPLFDPP 1623
Query: 659 RPGVKESVQVCRSAGIMVRMVTGDNINTAKAIARECGILTE--DGLAIEGPDFREKTQEE 716
R E+++ G+ V+M+TGD + K R G+ T ++ G + +
Sbjct: 1624RHDSAETIRRALDLGVSVKMITGDQLAIGKETGRRLGMGTNMYPSSSLLGDNKDQLGAVS 1803
Query: 717 MFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAG 776
+ +LI A P K+ +VK+L+ + +TGDG NDAPAL ADIG+A+
Sbjct: 1804IDDLIENADGFAGVFPEHKYEIVKRLQAR-KHICGMTGDGVNDAPALKIADIGIAVA-DS 1977
Query: 777 TEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVALL 826
T+ A+ ++D+++ + S I++ R+++ ++ + + +++ + +L
Sbjct: 1978TDAARSASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVL 2127
>BF004483 GP|21314227| type IIB calcium ATPase MCA5 {Medicago truncatula},
partial (11%)
Length = 551
Score = 176 bits (446), Expect = 4e-44
Identities = 83/109 (76%), Positives = 98/109 (89%)
Frame = +1
Query: 828 NFSSAVLTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKRAPLGRKGDFINSIMW 887
N + +G+APLTAVQLLWVNMIMDTLGALALATEPP DDLMKRAP+GRKG+FI+++MW
Sbjct: 46 NIICILSSGTAPLTAVQLLWVNMIMDTLGALALATEPPNDDLMKRAPVGRKGNFISNVMW 225
Query: 888 RNILGQALYQFVVIWFLQTVGKWVFFLRGPNAGVVLNTLIFNSFVFCQV 936
RNILGQ+LYQF+VIWFLQ+ GK +F L GPN+ +VLNTLIFN+FVFCQV
Sbjct: 226 RNILGQSLYQFMVIWFLQSKGKTIFSLDGPNSDLVLNTLIFNAFVFCQV 372
Score = 29.6 bits (65), Expect = 6.1
Identities = 12/15 (80%), Positives = 15/15 (100%)
Frame = +2
Query: 935 QVFNEINSREMEEVD 949
QVFNEINSREME+++
Sbjct: 506 QVFNEINSREMEKIN 550
>TC77796 H+-ATPase
Length = 3416
Score = 121 bits (303), Expect(2) = 3e-43
Identities = 103/375 (27%), Positives = 173/375 (45%), Gaps = 4/375 (1%)
Frame = +1
Query: 452 ICSDKTGTLTTNHMTVVKTCICMSSKEVNNKEHGLCSELPDSAQKLLLQSIFNNTGGEVV 511
+CSDKTGTLT N ++V K I + +K V K++ ++
Sbjct: 1075 LCSDKTGTLTLNKLSVEKNLIEVFAKGVE-KDY------------------------VIL 1179
Query: 512 VNKRGKREILGTPTESAILEFGLSLGGDPQKERQACKLVKVEPFNSQKKRMGVVVELPEG 571
+ R R T + AI + + DP++ R + V PFN KR + +G
Sbjct: 1180 LAARASR----TENQDAIDAAIVGMLADPKEARAGVREVHFFPFNPVDKRTALTYIDADG 1347
Query: 572 GLRAHCKGASEIVLAACDNVIDSKGDVVPLNAESRNYLESTIDQFAGEALRTLCLAYIEL 631
KGA E +L C+ K DV R STID+FA LR+L +A E+
Sbjct: 1348 NWHRSSKGAPEQILNLCN----CKEDV-------RKKAHSTIDKFAERGLRSLGVARQEI 1494
Query: 632 EHGFSAEDPIPASGYTCIGVVGIKDPVRPGVKESVQVCRSAGIMVRMVTGDNINTAKAIA 691
+ P + + +G++ + DP R E++ + G+ V+M+TGD + AK
Sbjct: 1495 PE---KDKDSPGAPWQFVGLLPLFDPPRHDSAETITRALNLGVNVKMITGDQLAIAKETG 1665
Query: 692 RECGILTE----DGLAIEGPDFREKTQEEMFELIPKIQVMARSSPLDKHTLVKQLRTTFG 747
R G+ T L + D + ELI K A P K+ +VK+L+
Sbjct: 1666 RRLGMGTNMYPSSSLLGQSKDAAVSALP-VDELIEKADGFAGVFPEHKYEIVKKLQER-K 1839
Query: 748 EVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVY 807
+ +TGDG NDAPAL ADIG+A+ A T+ A+ ++D+++ + S I++ R+++
Sbjct: 1840 HICGMTGDGVNDAPALKRADIGIAVADA-TDAARGASDIVLTEPGLSVIISAVLTSRAIF 2016
Query: 808 INIQKFVQFQLTVNV 822
++ + + +++ +
Sbjct: 2017 QRMKNYTIYAVSITI 2061
Score = 73.6 bits (179), Expect(2) = 3e-43
Identities = 76/332 (22%), Positives = 145/332 (42%), Gaps = 5/332 (1%)
Frame = +3
Query: 120 VSGIAEKLSTSTTKGLSGDSEARRIRQEVYGINKFAESEVRSFWIFVYEALQDMTLMILA 179
V + E+L S GL+ D A R+ +V+G NK E F F+ ++ ++ A
Sbjct: 153 VEEVFEQLKCSRA-GLTSDEGANRL--QVFGPNKLEEKRESKFLKFLGFMWNPLSWVMEA 323
Query: 180 VCAFVSLIVGIATEGWPQGSHDGLGIVASILLVVFVTATSDYRQSLQFKDLDKEKKKISI 239
A + I G P D +GI++ +++ ++ + L
Sbjct: 324 --AAIMAIALANGSGRPPDWQDFVGIISLLVINSTISFIEENNAGNAAAALMAGLAP-KT 494
Query: 240 QVTRNGYRQKMSIYNLLPGDLVHLSIGDQVPTDGLFVSGFSVLIDESSLTGESEPVMVTS 299
+V R+G + L+PGD++ + +GD +P D + G ++ +D+S+LTGES P +
Sbjct: 495 RVLRDGRWSEEDAAILVPGDIISIKLGDIIPADARLLEGDALSVDQSALTGESLP---AT 665
Query: 300 QNPF--LLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVATLIG 357
+NP + SG+ V+ G +V G+ T +GK + D T V T IG
Sbjct: 666 KNPSDEVFSGSTVKKGEIEAVVIATGVHTFFGKAAHLV-----DSTNQVGHFQKVLTAIG 830
Query: 358 K---VGLFFAVVTFVVLVKGLMSRKIREGRFWWWSADDAMEMLEFFAIAVTIVVVAVPEG 414
+ ++ +V++ + RK R+G D+ + +L + +P
Sbjct: 831 NFCICSIAVGILIELVVMYPIQHRKYRDG------IDNLLVLL----------IGGIPIA 962
Query: 415 LPLAVTLSLAFAMKKMMNDKALVRHLAACETM 446
+P +++++A ++ A+ + + A E M
Sbjct: 963 MPTVLSVTMAIGSHRLSQQGAITKRMTAIEEM 1058
>BG647796 GP|21314227|g type IIB calcium ATPase MCA5 {Medicago truncatula},
partial (9%)
Length = 780
Score = 156 bits (394), Expect = 4e-38
Identities = 74/97 (76%), Positives = 87/97 (89%)
Frame = +1
Query: 918 NAGVVLNTLIFNSFVFCQVFNEINSREMEEVDVFKGIWDNHVFVAVIGCTVVFQIIIVEY 977
++ +VLNTLIFN+FVFCQVFNEINSREME+++VFKGI DN+VFV VI T+ FQIIIVEY
Sbjct: 1 DSDLVLNTLIFNAFVFCQVFNEINSREMEKINVFKGILDNYVFVGVISATIFFQIIIVEY 180
Query: 978 LGTFANTTPLSLVQWIFCLSVGYVGMPIATYLKQIPV 1014
LGTFANTTPL+LVQW FCL VG++GMPIA LK+IPV
Sbjct: 181 LGTFANTTPLTLVQWFFCLFVGFMGMPIAARLKKIPV 291
>TC79651 similar to PIR|T52581|T52581 Ca2+-transporting ATPase (EC 3.6.1.38)
ECA3 [imported] - Arabidopsis thaliana, partial (38%)
Length = 1546
Score = 138 bits (347), Expect = 1e-32
Identities = 106/362 (29%), Positives = 181/362 (49%), Gaps = 37/362 (10%)
Frame = +3
Query: 681 GDNINTAKAIARECG----ILTEDGLAIEGPDFREKTQEEMFELIPKIQVMARSSPLDKH 736
G+N +TA+++ R+ G ++ + +F E + + ++ + R P K
Sbjct: 18 GNNKSTAESLCRKIGAFDHLIDFTEHSYTASEFEELPALQQTIALQRMALFTRVEPSHKR 197
Query: 737 TLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTI 796
LV+ L+ EVVA+TGDG NDAPAL +ADIG+AMG +GT VAK ++D+++ DDNF++I
Sbjct: 198 MLVEALQHQ-NEVVAMTGDGVNDAPALKKADIGIAMG-SGTAVAKSASDMVLADDNFASI 371
Query: 797 VTVAKWGRSVYINIQKFVQFQLTVNVVALLVNFSSAVLTGSAPLTAVQLLWVNMIMDTLG 856
V GR++Y N ++F+++ ++ N+ ++ F +AVL L VQLLWVN++ D L
Sbjct: 372 VAAVAEGRAIYNNTKQFIRYMISSNIGEVVCIFVAAVLGIPDTLAPVQLLWVNLVTDGLP 551
Query: 857 ALALATEPPTDDLMKRAPLGRKGDFINSIMWRNILGQALYQFVVIWF---LQTVGKWVFF 913
A A+ D+MK P RK +N + + G ++++VI L TV ++++
Sbjct: 552 ATAIGFNKQDSDVMKVKP--RK---VNEAV---VTGWLFFRYLVIGAYVGLATVAGFIWW 707
Query: 914 LRGPNAGVVL----------------------------NTLIFNSFVFCQVFNEINSREM 945
++G L +T+ V ++FN +N+
Sbjct: 708 FVYADSGPKLPYTELMNFDTCPTRETTYPCSIFEDRHPSTVAMTVLVVVEMFNALNNLSE 887
Query: 946 EEVDVFKGIWDNHVFVAVIGCTVVFQIII--VEYLGTFANTTPLSLVQWIFCLSVGYVGM 1003
+ + W N V I T++ I+I V L + TPLS W ++V Y+ +
Sbjct: 888 NQSLLVIPPWSNLWLVGSIVLTMLLHILILYVRPLSVLFSVTPLSWADW---MAVLYLSL 1058
Query: 1004 PI 1005
P+
Sbjct: 1059 PV 1064
>CB893893 similar to GP|8843813|db Ca2+-transporting ATPase-like protein
{Arabidopsis thaliana}, partial (11%)
Length = 645
Score = 122 bits (305), Expect(2) = 2e-31
Identities = 60/105 (57%), Positives = 83/105 (78%)
Frame = +1
Query: 243 RNGYRQKMSIYNLLPGDLVHLSIGDQVPTDGLFVSGFSVLIDESSLTGESEPVMVTSQNP 302
R G R ++SIY+L+ GD++ L+IG+QVP DG+ ++G S+ IDESS+TGES+ V S++P
Sbjct: 4 RGGRRVEISIYDLVVGDVIPLNIGNQVPADGVVITGHSLSIDESSMTGESKIVHKDSKDP 183
Query: 303 FLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQV 347
F++SG KV DGS TMLVT VG+ T+WG LMA++SE +ETPLQV
Sbjct: 184 FMMSGCKVADGSGTMLVTGVGINTEWGLLMASISEDTGEETPLQV 318
Score = 33.1 bits (74), Expect(2) = 2e-31
Identities = 16/26 (61%), Positives = 20/26 (76%)
Frame = +2
Query: 347 VKLNGVATLIGKVGLFFAVVTFVVLV 372
V+LNGVAT IG VGL AV+ +VL+
Sbjct: 395 VRLNGVATFIGIVGLSVAVLVLIVLL 472
>TC86961 similar to GP|1742951|emb|CAA70946.1 Ca2+-ATPase {Arabidopsis
thaliana}, partial (77%)
Length = 1488
Score = 129 bits (324), Expect = 6e-30
Identities = 75/178 (42%), Positives = 110/178 (61%), Gaps = 4/178 (2%)
Frame = +1
Query: 752 VTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQ 811
+TGDG NDAPAL ADIG+AMGIAGTEVAKE++D+++ DDNFS+IV GRS+Y N++
Sbjct: 55 MTGDGVNDAPALKLADIGIAMGIAGTEVAKEASDMVLADDNFSSIVAAVGEGRSIYNNMK 234
Query: 812 KFVQFQLTVNVVALLVNFSSAVLTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMK 871
F+++ ++ N+ + F +A L L VQLLWVN++ D A AL PP D+MK
Sbjct: 235 AFIRYMISSNIGEVASIFLTAALGIPEGLIPVQLLWVNLVTDGPPATALGFNPPDKDIMK 414
Query: 872 RAPLGRKGDFINSIMWRNILGQALYQFVVIWF---LQTVGKW-VFFLRGPNAGVVLNT 925
+ P IN +W L++++VI L TVG + +++ G G+ L++
Sbjct: 415 KPPRRSDDSLIN--LW------ILFRYLVIGIYVGLATVGVFIIWYTHGSFMGIDLSS 564
>CB892538 similar to SP|Q9XES1|ECA Calcium-transporting ATPase 4 endoplasmic
reticulum-type (EC 3.6.3.8). [Mouse-ear cress], partial
(24%)
Length = 805
Score = 80.5 bits (197), Expect(2) = 1e-25
Identities = 60/185 (32%), Positives = 97/185 (52%), Gaps = 22/185 (11%)
Frame = +2
Query: 555 FNSQKKRMGVVVELPEGGLRAHC-KGASEIVLAACDNVIDSKGDVVPLNAESRNYLESTI 613
F+ +K MGV+V+ G ++ KGA E VL V G VV L+ ++N + +
Sbjct: 59 FDRDRKSMGVIVDSGVGKKKSLLVKGAVENVLDRSSKVQLRDGSVVKLDNNAKNLILQAL 238
Query: 614 DQFAGEALRTLCLAYI-ELEH--GFSAEDPIPA-------SGYTCI-------GVVGIKD 656
+ + ALR L AY EL + ++ + PA + Y+ I G+VG++D
Sbjct: 239 HEMSTSALRCLGFAYKDELTNFENYNGNEDHPAHQLLLDPNNYSSIEDELIFVGLVGLRD 418
Query: 657 PVRPGVKESVQVCRSAGIMVRMVTGDNINTAKAIARECGILTED----GLAIEGPDFREK 712
P R V ++++ CR+AGI V ++TGDN NTA+AI RE G+ + ++ G DF E
Sbjct: 419 PPREEVYQAIEDCRAAGIRVMVITGDNKNTAEAICREIGVFAPNENISSKSLTGKDFMEL 598
Query: 713 TQEEM 717
+++
Sbjct: 599 RDKKL 613
Score = 55.1 bits (131), Expect(2) = 1e-25
Identities = 28/35 (80%), Positives = 30/35 (85%)
Frame = +3
Query: 747 GEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAK 781
GEVVA+TGDG NDA AL ADIG+AMGIA TEVAK
Sbjct: 699 GEVVAMTGDGCNDATALKLADIGIAMGIARTEVAK 803
>BQ138304 homologue to GP|6688833|emb putative calcium P-type ATPase
{Neurospora crassa}, partial (13%)
Length = 412
Score = 114 bits (284), Expect = 2e-25
Identities = 61/138 (44%), Positives = 91/138 (65%)
Frame = +3
Query: 714 QEEMFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMG 773
+ E E + +R+ P K LV L+ GEVVA+TGDG NDAPAL +ADIG+AMG
Sbjct: 3 EAEQMEAAKNASLFSRTEPTHKSKLVDLLQKA-GEVVAMTGDGVNDAPALKKADIGVAMG 179
Query: 774 IAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVALLVNFSSAV 833
+GT+VAK +AD++++DDNF+TI + GRS+Y N Q+F+++ ++ N+ ++ F +A
Sbjct: 180 -SGTDVAKLAADMVLVDDNFATIEGAVEEGRSIYNNTQQFIRYLISSNIGEVVSIFLTAA 356
Query: 834 LTGSAPLTAVQLLWVNMI 851
L VQLLWVN++
Sbjct: 357 AGMPEALIPVQLLWVNLV 410
>TC90329 GP|21314227|gb|AAM44081.1 type IIB calcium ATPase MCA5 {Medicago
truncatula}, partial (5%)
Length = 547
Score = 102 bits (255), Expect = 6e-22
Identities = 49/59 (83%), Positives = 53/59 (89%)
Frame = +1
Query: 1 MESYLNESFGGVKSKNSTEEALSKWRKVCGVVKNPKRRFRFTANLSKRYEAAAMRRTNQ 59
ME+YL E+FGGVKSKNS+EEAL +WR VCG VKNPKRRFRFTANL KR EAAAMRRTNQ
Sbjct: 94 MENYLQENFGGVKSKNSSEEALRRWRDVCGFVKNPKRRFRFTANLDKRGEAAAMRRTNQ 270
>CB893927 homologue to GP|17342714|g type IIA calcium ATPase {Medicago
truncatula}, partial (13%)
Length = 413
Score = 84.7 bits (208), Expect(2) = 4e-19
Identities = 48/116 (41%), Positives = 74/116 (63%), Gaps = 6/116 (5%)
Frame = +2
Query: 670 RSAGIMVRMVTGDNINTAKAIARECGILTED----GLAIEGPDFREKTQEEMFELIPKI- 724
+ AGI V ++TG+N +TA+AI +E + + D G ++ G +F + E +L+ +
Sbjct: 2 KQAGIRVMVITGENKSTAEAICKEIKLFSTDEDLTGQSLTGKEFMSLSHSEQVKLLLRNG 181
Query: 725 -QVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEV 779
+ +R+ P K +V+ L+ GE+VA+TGDG NDAPAL ADIG+AMGI GTE+
Sbjct: 182 GKGFSRAEPRHKQEIVRLLKE-MGEIVAMTGDGVNDAPALKLADIGMAMGITGTEM 346
Score = 29.3 bits (64), Expect(2) = 4e-19
Identities = 12/22 (54%), Positives = 17/22 (76%)
Frame = +1
Query: 790 DDNFSTIVTVAKWGRSVYINIQ 811
DDNFSTIV+ GR++Y N++
Sbjct: 343 DDNFSTIVSAIAEGRAIYNNMK 408
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,082,961
Number of Sequences: 36976
Number of extensions: 343924
Number of successful extensions: 1757
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 1686
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1717
length of query: 1014
length of database: 9,014,727
effective HSP length: 106
effective length of query: 908
effective length of database: 5,095,271
effective search space: 4626506068
effective search space used: 4626506068
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0347b.6


