

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0346.5
(350 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
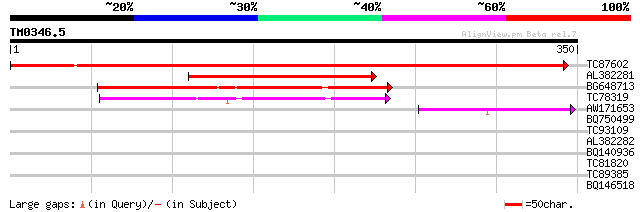
Score E
Sequences producing significant alignments: (bits) Value
TC87602 similar to GP|21741453|emb|CAD41397. OJ000223_09.9 {Oryz... 545 e-155
AL382281 similar to GP|6018193|gb| nucleotide-binding protein lo... 144 5e-35
BG648713 similar to GP|16924108|gb putative nucleotide-binding p... 132 2e-31
TC78319 homologue to GP|21592386|gb|AAM64337.1 mrp protein puta... 116 1e-26
AW171653 48 6e-06
BQ750499 similar to OMNI|NT01MC4764 transport protein HasD puta... 33 0.13
TC93109 similar to GP|5824321|emb|CAB54139.1 ATPase {Solanum tub... 33 0.13
AL382282 similar to GP|1360147|emb| Nbp35p {Saccharomyces cerevi... 32 0.37
BQ140936 similar to GP|21537349|gb| ATP binding protein-like {Ar... 32 0.48
TC81820 similar to GP|6056208|gb|AAF02825.1| putative ATPase {Ar... 28 5.3
TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resist... 28 6.9
BQ146518 weakly similar to PIR|C86226|C862 protein T31J12.4 [imp... 28 6.9
>TC87602 similar to GP|21741453|emb|CAD41397. OJ000223_09.9 {Oryza sativa},
partial (95%)
Length = 1263
Score = 545 bits (1404), Expect = e-155
Identities = 264/345 (76%), Positives = 301/345 (86%)
Frame = +2
Query: 1 MEKGDIPENANDHCPGSQSDSAGKSDGCQGCPNQQTCATSSPKGPDSDLVAIKERMATVK 60
ME GDIPE+AN+HCPG QSDSAGKSD C+GCPNQQ CAT+ PKGPD D+VAI ERMATVK
Sbjct: 80 MENGDIPEDANEHCPGPQSDSAGKSDACEGCPNQQICATA-PKGPDPDMVAIAERMATVK 256
Query: 61 HKILVLSGKGGVGKSTFSAQLAFALAAKDFEVGLLDIDICGPSIPKMLGLEGHELHRSNF 120
HKILVLSGKGGVGKSTFSAQLAFALAA+DF+VGLLDIDICGPSIPKMLGLEG E+H+SN
Sbjct: 257 HKILVLSGKGGVGKSTFSAQLAFALAARDFQVGLLDIDICGPSIPKMLGLEGQEIHQSNL 436
Query: 121 GCDPVYVQPNLGVMSIGFVNSDPDDPIILRGPRKSWLITEFLKDVYWNELDFLIVDAPPG 180
G PVYV+ NLGVMSIGF+ PD+ +I RGPRK+ LI +FLKDVYW ELDFLIVDAPPG
Sbjct: 437 GWSPVYVESNLGVMSIGFMLPHPDEAVIWRGPRKNGLIKQFLKDVYWGELDFLIVDAPPG 616
Query: 181 TSDEQISIVKLLGATGLDGAIIITTPQQVSLTDVRKGVNFCKQAEVKVLGVVENMSGLCQ 240
TSDE ISIV+ L A +DGAII+TTPQQVSL DV+K VNFCK+ VKVLGVVENMSGL Q
Sbjct: 617 TSDEHISIVQCLDAANVDGAIIVTTPQQVSLIDVKKEVNFCKKVGVKVLGVVENMSGLSQ 796
Query: 241 PVMDFKFLKLASNGEQKDVTEWFLKFMREKAPEMVDMIACTEVFDSSGGGAVRMCKEMMI 300
P+ + KF+K+ NGE KDVTEW ++M+EKAPEM+++IAC+EVFDSS GGA++MC EM +
Sbjct: 797 PISNLKFMKITDNGEMKDVTEWISEYMKEKAPEMLNLIACSEVFDSSRGGALKMCNEMAV 976
Query: 301 PFLGKVPLDPKLCKAAEEGRSCFDDKDCVASAFALKNIIEKLMET 345
PFLGKVPLDP+LCKAAEEGRSCF DKDCV SA AL+ II+KLMET
Sbjct: 977 PFLGKVPLDPQLCKAAEEGRSCFADKDCVVSAPALQKIIDKLMET 1111
>AL382281 similar to GP|6018193|gb| nucleotide-binding protein long form {Mus
musculus}, partial (35%)
Length = 350
Score = 144 bits (363), Expect = 5e-35
Identities = 66/116 (56%), Positives = 91/116 (77%)
Frame = +1
Query: 111 EGHELHRSNFGCDPVYVQPNLGVMSIGFVNSDPDDPIILRGPRKSWLITEFLKDVYWNEL 170
EG ++H+S G PVYVQ NLGVMSIGF+ S+PDD +I RGP K+ LI +FLKDV W ++
Sbjct: 1 EGEQIHQSLSGWSPVYVQENLGVMSIGFMLSNPDDAVIWRGPXKNGLIKQFLKDVDWGDI 180
Query: 171 DFLIVDAPPGTSDEQISIVKLLGATGLDGAIIITTPQQVSLTDVRKGVNFCKQAEV 226
D+++VD PPGTSDE +SI + L + +DGAIIITTP +V+L DVRK ++FC++ ++
Sbjct: 181 DYMLVDTPPGTSDEHLSIAQYLKESVIDGAIIITTPXEVALLDVRKEIDFCRKVKI 348
>BG648713 similar to GP|16924108|gb putative nucleotide-binding protein
{Oryza sativa}, partial (69%)
Length = 754
Score = 132 bits (332), Expect = 2e-31
Identities = 77/183 (42%), Positives = 112/183 (61%), Gaps = 1/183 (0%)
Frame = +1
Query: 55 RMATVKHKILVLSGKGGVGKSTFSAQLAFALAAK-DFEVGLLDIDICGPSIPKMLGLEGH 113
R+ VK I + SGKGGVGKST + LA ALA+K +VGLLD D+ GP+IP M+ ++
Sbjct: 136 RIDGVKDTIAIASGKGGVGKSTTAVNLAVALASKFQLKVGLLDADVYGPNIPIMMNIDTK 315
Query: 114 ELHRSNFGCDPVYVQPNLGVMSIGFVNSDPDDPIILRGPRKSWLITEFLKDVYWNELDFL 173
+ P+ + MSIGF+ + + PI+ RGP S + + + V W LD L
Sbjct: 316 PEVTQDNKMIPIDSY-GIKCMSIGFL-VEKNAPIVWRGPMVSKALEKMTRGVDWGNLDIL 489
Query: 174 IVDAPPGTSDEQISIVKLLGATGLDGAIIITTPQQVSLTDVRKGVNFCKQAEVKVLGVVE 233
++D PPGT D QIS+ + L L GA+I++TPQ V+L D R+GV + ++ +LG++E
Sbjct: 490 VIDMPPGTGDVQISMSQNLQ---LSGALIVSTPQDVALMDARRGVQMFNKVDIPILGIIE 660
Query: 234 NMS 236
NMS
Sbjct: 661 NMS 669
>TC78319 homologue to GP|21592386|gb|AAM64337.1 mrp protein putative
{Arabidopsis thaliana}, partial (60%)
Length = 1243
Score = 116 bits (291), Expect = 1e-26
Identities = 71/182 (39%), Positives = 101/182 (55%), Gaps = 2/182 (1%)
Frame = +2
Query: 56 MATVKHKILVLSGKGGVGKSTFSAQLAFALAAKDFEVGLLDIDICGPSIPKMLGLEGHEL 115
+ T+ + I V S KGGVGKST + LA+ LA VG+ D DI GPS+P M+ E L
Sbjct: 545 LQTISNIIAVSSCKGGVGKSTVAVNLAYTLADMGARVGIFDADIYGPSLPTMVSPENRIL 724
Query: 116 HRSNFGCDPVYVQPNLGV--MSIGFVNSDPDDPIILRGPRKSWLITEFLKDVYWNELDFL 173
N + +GV +S GF I+RGP S + + L W ELD+L
Sbjct: 725 -EMNPEKKTIIPTEYMGVKLVSFGFAG---QGRAIMRGPMVSGVTNQLLTTTEWGELDYL 892
Query: 174 IVDAPPGTSDEQISIVKLLGATGLDGAIIITTPQQVSLTDVRKGVNFCKQAEVKVLGVVE 233
++D PPGT D Q+++ +++ T A+I+TTPQ++S DV KGV + +V + VVE
Sbjct: 893 VIDMPPGTGDIQLTLCQIVPLT---AAVIVTTPQKLSFIDVAKGVRMFSKLKVPCVAVVE 1063
Query: 234 NM 235
NM
Sbjct: 1064NM 1069
>AW171653
Length = 390
Score = 47.8 bits (112), Expect = 6e-06
Identities = 24/104 (23%), Positives = 51/104 (48%), Gaps = 7/104 (6%)
Frame = +1
Query: 253 NGEQKDVTEWFLKFMREKAPEMVDMIACTEVFDSSGGGAVR-------MCKEMMIPFLGK 305
N E DV+ + ++ PE+++ + +F S + M +P+LG+
Sbjct: 16 NSEGVDVSSEVIAKLQASCPEVLNYSLHSNLFTPSAASSSAAANTPKAMADRFNVPYLGR 195
Query: 306 VPLDPKLCKAAEEGRSCFDDKDCVASAFALKNIIEKLMETVESV 349
+P+DP + A EEG+S ++ ++ ++I+ K++E S+
Sbjct: 196 LPMDPNMLNACEEGKSFLEEYPVSSATSEFRSIVSKIVEATSSL 327
>BQ750499 similar to OMNI|NT01MC4764 transport protein HasD putative
{Magnetococcus sp. MC-1}, partial (3%)
Length = 800
Score = 33.5 bits (75), Expect = 0.13
Identities = 22/71 (30%), Positives = 35/71 (48%), Gaps = 5/71 (7%)
Frame = +1
Query: 18 QSDSAGKSDGCQ-----GCPNQQTCATSSPKGPDSDLVAIKERMATVKHKILVLSGKGGV 72
+ ++ G+ DG Q G P + C G D L +K ++ +LVL+G GG
Sbjct: 448 RKENLGQFDGSQIRGLCGAPEEPQCM-----GVDEPLNKLKIQLLKDGVSVLVLTGLGGS 612
Query: 73 GKSTFSAQLAF 83
GKST + +L +
Sbjct: 613 GKSTLAKKLCW 645
>TC93109 similar to GP|5824321|emb|CAB54139.1 ATPase {Solanum tuberosum},
partial (44%)
Length = 743
Score = 33.5 bits (75), Expect = 0.13
Identities = 17/55 (30%), Positives = 27/55 (48%)
Frame = +3
Query: 32 PNQQTCATSSPKGPDSDLVAIKERMATVKHKILVLSGKGGVGKSTFSAQLAFALA 86
P ++ S P + + +A + K +L GKGGVGK++ +A LA A
Sbjct: 183 PPTKSFQVKSVAAPTESISVFDDMVAGTERKYYMLGGKGGVGKTSCAASLAVKFA 347
>AL382282 similar to GP|1360147|emb| Nbp35p {Saccharomyces cerevisiae},
partial (8%)
Length = 260
Score = 32.0 bits (71), Expect = 0.37
Identities = 15/40 (37%), Positives = 24/40 (59%)
Frame = +1
Query: 303 LGKVPLDPKLCKAAEEGRSCFDDKDCVASAFALKNIIEKL 342
LG +PLDP++ KA + G S D+ + A ++II K+
Sbjct: 1 LGSIPLDPRIGKACDTGVSFLDEYSDSPACKAFQDIIAKI 120
>BQ140936 similar to GP|21537349|gb| ATP binding protein-like {Arabidopsis
thaliana}, partial (7%)
Length = 270
Score = 31.6 bits (70), Expect = 0.48
Identities = 15/29 (51%), Positives = 20/29 (68%)
Frame = +1
Query: 55 RMATVKHKILVLSGKGGVGKSTFSAQLAF 83
R+ VK I + SGKGGVGKST + + +F
Sbjct: 160 RIDGVKDTIAIASGKGGVGKSTXAGKNSF 246
>TC81820 similar to GP|6056208|gb|AAF02825.1| putative ATPase {Arabidopsis
thaliana}, partial (69%)
Length = 1320
Score = 28.1 bits (61), Expect = 5.3
Identities = 13/33 (39%), Positives = 20/33 (60%)
Frame = +2
Query: 54 ERMATVKHKILVLSGKGGVGKSTFSAQLAFALA 86
E + + + +L GKGGVGK++ +A LA A
Sbjct: 83 EMVRDKERRYYMLGGKGGVGKTSCAASLAIKFA 181
>TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resistance gene
analog protein {Medicago ruthenica}, partial (98%)
Length = 1285
Score = 27.7 bits (60), Expect = 6.9
Identities = 25/90 (27%), Positives = 37/90 (40%), Gaps = 22/90 (24%)
Frame = +3
Query: 48 DLVAIKERMATVKHKILVLS----------GKGGVGKST------------FSAQLAFAL 85
DLV I R +KH++L+ S G GG GK+T F A
Sbjct: 591 DLVGINSRTEALKHQLLLNSVDGVRVIGIWGMGGKGKTTLAMNLYGQICHRFDASCFIDD 770
Query: 86 AAKDFEVGLLDIDICGPSIPKMLGLEGHEL 115
+K F + ID + + LG+E H++
Sbjct: 771 VSKIFRLHDGPIDAQKQILHQTLGIEHHQI 860
>BQ146518 weakly similar to PIR|C86226|C862 protein T31J12.4 [imported] -
Arabidopsis thaliana, partial (11%)
Length = 432
Score = 27.7 bits (60), Expect = 6.9
Identities = 12/34 (35%), Positives = 19/34 (55%)
Frame = -2
Query: 120 FGCDPVYVQPNLGVMSIGFVNSDPDDPIILRGPR 153
F PV +QP LG++ + F+ P+ IL P+
Sbjct: 194 FAIHPVLMQPQLGILKLNFLIGVPEKNNILGVPK 93
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,460,934
Number of Sequences: 36976
Number of extensions: 135348
Number of successful extensions: 600
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 593
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 594
length of query: 350
length of database: 9,014,727
effective HSP length: 97
effective length of query: 253
effective length of database: 5,428,055
effective search space: 1373297915
effective search space used: 1373297915
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0346.5


