

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0346.14
(1622 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
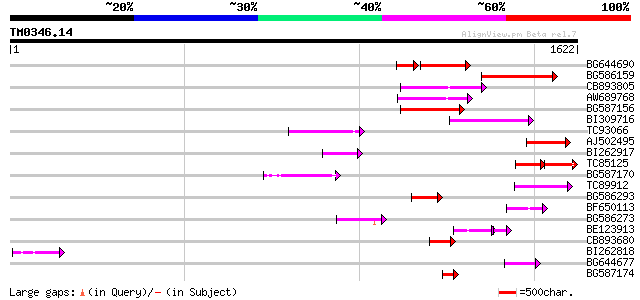
Score E
Sequences producing significant alignments: (bits) Value
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 188 9e-65
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 206 6e-53
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 171 2e-42
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 169 6e-42
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 163 6e-40
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 152 1e-36
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 140 3e-33
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 119 1e-26
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 117 4e-26
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 70 8e-26
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 112 2e-24
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 107 3e-23
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 96 1e-19
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 87 5e-17
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 81 3e-15
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 68 1e-14
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 72 2e-12
BI262818 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 69 1e-11
BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T... 63 8e-10
BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse ... 59 2e-08
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 188 bits (477), Expect(2) = 9e-65
Identities = 89/141 (63%), Positives = 114/141 (80%)
Frame = -2
Query: 1176 VVRNKARLVAKGYSQQEGIDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNG 1235
+ RNK++LV +GY+Q+EGIDY E F PVAR+EAIR+LI+F+ L+QMDVKSAF+NG
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 1236 YISEEVYVHQPPGFEDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDT 1295
+ EEV+V QPPGFED + P+HVF+L K+LYGLKQAPRAWYERLS FLL+N F RGK+D
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 1296 TLFCKTYKDDILIVQIYVDKI 1316
TLF + ++LI+Q+YVD I
Sbjct: 64 TLFLLKRE*ELLIIQVYVDDI 2
Score = 79.3 bits (194), Expect(2) = 9e-65
Identities = 34/64 (53%), Positives = 49/64 (76%)
Frame = -3
Query: 1107 LLSMKGLVSLIEPKSIDEALQDKDWILAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVF 1166
L+S +S IEPK++ EAL+D DWI +M+EE+++F ++ VW LV +P+ VIGT+WVF
Sbjct: 621 LVSFSAFISSIEPKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKTVIGTRWVF 442
Query: 1167 RNKL 1170
RNKL
Sbjct: 441 RNKL 430
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 206 bits (524), Expect = 6e-53
Identities = 103/217 (47%), Positives = 142/217 (64%), Gaps = 1/217 (0%)
Frame = +1
Query: 1351 IQVDQTPEGTYIHQSKYTKELLKKFNMTESTIAKTPMHPTCILEKEDASGKVCQKLYRCM 1410
++V Q EG YI Q KY +LL++F M +S +++ P+ P C L K++ KV Y+ +
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 1411 IGSLLYLTASRPDNLFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYK 1470
+G L+YL A+RPD ++ + L +RF + P E H+ AVKR+LRYL GT NLG+MYK+ K
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 1471 LSGYCDVDYAGDRT*RKSTFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISAAICSTQM 1530
L Y D DYAGD RKST G L S VSW+SK+Q + LST KAE+I+AA C+ Q
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQS 540
Query: 1531 LWMKHQLEDYQILES-NIPIYCDNTAAISLSKNPILH 1566
+WM+ LE +S +I +YCDN + I LSKNP+LH
Sbjct: 541 VWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 171 bits (433), Expect = 2e-42
Identities = 90/249 (36%), Positives = 147/249 (58%), Gaps = 4/249 (1%)
Frame = +3
Query: 1118 EPKSIDEALQDKDWILAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVV 1177
+P + +EA++ + W +M E+ +N+ W L IG KW+F+ KLNE G++
Sbjct: 39 DPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNENGEIE 218
Query: 1178 RNKARLVAKGYSQQEGIDYTETFVPVARLEAIRLLISFSV---NHNIVLHQMDVKSAFLN 1234
+ KARLVAKGYSQQ G+DYTE F PVAR + IR++I+ + + + M + +
Sbjct: 219 KYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAAQIKRDGVCIS*M*KAHSCME 398
Query: 1235 GYISEEVYVHQPPGFEDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVD 1294
+ + + ++ + ++K++LYGLKQAPRAWY R+ ++ + F + +
Sbjct: 399 N*MRKFLLINHRVM*RRVIS----*RVKRALYGLKQAPRAWYSRIEAYFTKEGFEKCPYE 566
Query: 1295 TTLFCKTYK-DDILIVQIYVDKIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQV 1353
TLF K + ILI+ +YVD +IF ++++ +EF K M+ EF MS +G++ YFLG++V
Sbjct: 567 HTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGKMHYFLGVEV 746
Query: 1354 DQTPEGTYI 1362
Q +G YI
Sbjct: 747 TQNEKGIYI 773
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 169 bits (429), Expect = 6e-42
Identities = 95/218 (43%), Positives = 126/218 (57%), Gaps = 3/218 (1%)
Frame = +1
Query: 1110 MKGLVSLIEPKSID---EALQDKDWILAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVF 1166
+K V L EP D L D W+ AM+ E N W LV P IG KWV+
Sbjct: 25 LKPKVFLTEPCPQDCENLPLSDPRWLQAMKTEYKALIDNKTWDLVPLPPHKKAIGCKWVY 204
Query: 1167 RNKLNEKGDVVRNKARLVAKGYSQQEGIDYTETFVPVARLEAIRLLISFSVNHNIVLHQM 1226
R K N G V + KARLVAKG+SQ G DYTETF PV + IRL+++ ++ + + Q+
Sbjct: 205 RVKENPDGSVNKFKARLVAKGFSQTLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQI 384
Query: 1227 DVKSAFLNGYISEEVYVHQPPGFEDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLEN 1286
D+ +AFLNG++ EEVY+ QP GFE N V KL KSLYGLKQAPRAWYE L+S ++
Sbjct: 385 DINNAFLNGFLQEEVYMSQPQGFE-AANKSLVCKLNKSLYGLKQAPRAWYEXLTSAQIQF 561
Query: 1287 EFVRGKVDTTLFCKTYKDDILIVQIYVDKIIFGSANQS 1324
F + + D +L + + IYVD I+ ++ S
Sbjct: 562 GFTKSRCDPSLLIYNQNGACIYLXIYVDDILITGSSAS 675
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 163 bits (412), Expect = 6e-40
Identities = 79/183 (43%), Positives = 116/183 (63%)
Frame = -1
Query: 1119 PKSIDEALQDKDWILAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVR 1178
P+S +EA++DK+W ++ E KND W + P+ + ++W+F K G + R
Sbjct: 558 PRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSSRWIFTIKYKADGSIER 379
Query: 1179 NKARLVAKGYSQQEGIDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYIS 1238
K RLVA+G++ G DY ETF PVA+L IR+++S +VN L QMDVK+AFL G +
Sbjct: 378 KKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQMDVKNAFLQGELE 199
Query: 1239 EEVYVHQPPGFEDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLF 1298
+EVY++ PPG E +V +LKK++YGLKQ+PRAWY +LS+ L F + ++D TLF
Sbjct: 198 DEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLNGRGFRKSELDHTLF 19
Query: 1299 CKT 1301
T
Sbjct: 18 TLT 10
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 152 bits (383), Expect = 1e-36
Identities = 88/241 (36%), Positives = 130/241 (53%)
Frame = +2
Query: 1258 VFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDKII 1317
V +L+KS+YGLKQA R WY +LS L+ +++ D +LF K + +YVD I+
Sbjct: 20 VCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDIV 199
Query: 1318 FGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNM 1377
+ S + + F++ +G L+YFLG++V ++ +G ++Q KYT ELL+
Sbjct: 200 LAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDSGN 379
Query: 1378 TESTIAKTPMHPTCILEKEDASGKVCQKLYRCMIGSLLYLTASRPDNLFSVHLCARFQSD 1437
TP + L D+ + YR +IG L+YLT +RPD F+V ++F S
Sbjct: 380 LAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFVSK 559
Query: 1438 PRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAGDRT*RKSTFGNCQFLG 1497
P++ H A R+L+YLK GL Y TS KLS + D D+A T RKS G FLG
Sbjct: 560 PQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGYWVFLG 739
Query: 1498 S 1498
S
Sbjct: 740 S 742
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 140 bits (354), Expect = 3e-33
Identities = 77/217 (35%), Positives = 116/217 (52%), Gaps = 2/217 (0%)
Frame = +1
Query: 799 LELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHSVFSTFIAQVQNEK 858
L+ +H DL+GP K S GG+RY M I+DD+ R WV FL K+E+ F + V+ +
Sbjct: 97 LDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILVETQT 276
Query: 859 ACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVERKNRTLQEMARTML 918
+ ++ +D+ EF + F ++GIA + PR PQQNGV ER RTL E AR ML
Sbjct: 277 GKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCML 456
Query: 919 QEIGM--AKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIKPNISYFHSFGCVCYVL 976
G+ + W E +TAC++ NR + K P ++W + S FGC Y L
Sbjct: 457 SNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYAL 636
Query: 977 NTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKT 1013
+L ++ +C+ L Y+ SKG+R + +D K+
Sbjct: 637 VNDGKL---APRAGECIFLSYASESKGYRLWCSDPKS 738
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 119 bits (297), Expect = 1e-26
Identities = 57/128 (44%), Positives = 88/128 (68%), Gaps = 2/128 (1%)
Frame = +2
Query: 1478 DYAGDRT*RKSTFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISAAICSTQMLWMKHQL 1537
D+AGD RKST G LG+ +SW+SK+Q + STA+AEYI++ C+TQ +W++ L
Sbjct: 5 DWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRIL 184
Query: 1538 EDYQILESNIP--IYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDT 1595
E E N P IYCDN +AI+LSKNP+ H R+KHI++++H IR+ + + +++++ T
Sbjct: 185 EVMHH-EQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 1596 DHKWADIY 1603
+ K ADI+
Sbjct: 362 EEKIADIF 385
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza sativa}
[Oryza sativa (japonica cultivar-group)], partial (8%)
Length = 426
Score = 117 bits (293), Expect = 4e-26
Identities = 58/113 (51%), Positives = 68/113 (59%)
Frame = +1
Query: 896 TPQQNGVVERKNRTLQEMARTMLQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYEL 955
TPQQNGV ER NRTL E R ML+ GMAK FWAE V TACY+ NR I KTP E+
Sbjct: 85 TPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACYVINRSPSTVIDLKTPMEM 264
Query: 956 WKNIKPNISYFHSFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYN 1008
WK + S H FGC YV+ K D KS KC+ LGY+D KG+ ++
Sbjct: 265 WKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYADNVKGYXLWD 423
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 70.1 bits (170), Expect(2) = 8e-26
Identities = 38/87 (43%), Positives = 54/87 (61%)
Frame = +1
Query: 1446 VKRILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAGDRT*RKSTFGNCQFLGSNLVSWAS 1505
VKRI+RY+KGT+ + + + SE + GY D D+AGD RKST G L VSW S
Sbjct: 1 VKRIMRYIKGTSGVAVCFGG-SELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLS 177
Query: 1506 KRQSTIELSTAKAEYISAAICSTQMLW 1532
K Q+ + LST +AEY++A + + L+
Sbjct: 178 KLQTVVALSTTEAEYMAAYLKHARKLF 258
Score = 67.0 bits (162), Expect(2) = 8e-26
Identities = 31/95 (32%), Positives = 61/95 (63%), Gaps = 1/95 (1%)
Frame = +3
Query: 1529 QMLWMKHQLEDYQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVL 1588
+ +WM+ +E+ + I +YCD+ +A+ +++NP HSR KHI ++YHF+R+ V++G +
Sbjct: 249 EAIWMQRLMEELGHKQEQITVYCDSQSALHIARNPAFHSRTKHIGIQYHFVREVVEEGSV 428
Query: 1589 LLKFVDTDHKWAD-IYKALSRG*I*FHYEKSEYGL 1622
++ + T+ AD + K+++ F + +S YGL
Sbjct: 429 DMQKIHTNDNLADAMTKSINTD--KFIWCRSSYGL 527
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 112 bits (279), Expect = 2e-24
Identities = 80/226 (35%), Positives = 118/226 (51%), Gaps = 5/226 (2%)
Frame = -3
Query: 725 NVKCLL----SVNEEQ-WV*HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDALCEACQK 779
N KC S+N++ W H RLGH R LNL+ LP + F + CEAC
Sbjct: 632 NYKCSFTSSSSLNKDALW--HARLGHPHGRA------LNLM--LPGVVFENKN-CEACIL 486
Query: 780 GKFTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSR 839
GK K F + V + +L++ DL+ + S +Y + +D+ S++TW+ +
Sbjct: 485 GKHCKNVFPRTSTVYENC-FDLIYTDLW-TAPSLSRDNHKYFVTFIDEKSKYTWLTLIPS 312
Query: 840 KDESHSVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQ 899
KD F F A V N +I +RSD+GGE+ + F+S D +GI H SCP TPQQ
Sbjct: 311 KDRVIDAFKNFQAYVTNHYHAKIKILRSDNGGEYTSYAFKSHLDHHGILHQTSCPYTPQQ 132
Query: 900 NGVVERKNRTLQEMARTMLQEIGMAKHFWAEVVNTACYIQNRISVR 945
NGV +RKN+ L E+AR+++ + V+TACY+ N I +
Sbjct: 131 NGVAKRKNKHLMEVARSLMFQAN---------VSTACYLINWIPTK 21
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 107 bits (268), Expect = 3e-23
Identities = 54/166 (32%), Positives = 95/166 (56%), Gaps = 2/166 (1%)
Frame = +1
Query: 1445 AVKRILRYLKGTTNLGLMYKKTSEYK--LSGYCDVDYAGDRT*RKSTFGNCQFLGSNLVS 1502
A+K +L+YL + L Y K ++ + L GY D DYAG+ RKS G L +S
Sbjct: 4 ALKWVLKYLNESLKSSLKYTKAAQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTTIS 183
Query: 1503 WASKRQSTIELSTAKAEYISAAICSTQMLWMKHQLEDYQILESNIPIYCDNTAAISLSKN 1562
W + +QS + LST +AEYI+ +W+K + + I + + I+CD+ +AI L+ +
Sbjct: 184 WKANQQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGITQEYVKIHCDSQSAIHLANH 363
Query: 1563 PILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHKWADIYKALSR 1608
+ H R KHI+++ HFIRD ++ ++++ + ++ AD++ L +
Sbjct: 364 QVYHERTKHIDIRLHFIRDMIESKEIVVEKMASEENPADVFTKLKK 501
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 95.5 bits (236), Expect = 1e-19
Identities = 44/87 (50%), Positives = 62/87 (70%)
Frame = +2
Query: 1150 LVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAKGYSQQEGIDYTETFVPVARLEAI 1209
LVKKP V IG +W+++ K NE G +++ KARLVAKGY +Q+GID+ E F PV R+E I
Sbjct: 65 LVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPVVRIETI 244
Query: 1210 RLLISFSVNHNIVLHQMDVKSAFLNGY 1236
LL++ + + +H +DVK AFLNG+
Sbjct: 245 *LLLALAATNGC*IHHIDVKIAFLNGH 325
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 87.0 bits (214), Expect = 5e-17
Identities = 46/122 (37%), Positives = 70/122 (56%), Gaps = 3/122 (2%)
Frame = +1
Query: 1421 RPDNLFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMY---KKTSEYKLSGYCDV 1477
RPD +SV + ++F DPR+ HL A RILRY++GT GL++ K+ Y+L Y D
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICYSDS 300
Query: 1478 DYAGDRT*RKSTFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISAAICSTQMLWMKHQL 1537
D+ GD R+ST G +SW +K+Q LS+ +AEYI+ + Q LW+ +
Sbjct: 301 DWCGD---RRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSVI 471
Query: 1538 ED 1539
++
Sbjct: 472 KE 477
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 81.3 bits (199), Expect = 3e-15
Identities = 42/148 (28%), Positives = 81/148 (54%), Gaps = 6/148 (4%)
Frame = -2
Query: 935 ACYIQNRISVRRILNKTPYELWKNIKPNISYFHSFGCVCYVLNTKDRLHKFDAKSSKCLL 994
ACY+ NRI R + ++ P+E+ KP+++Y FGC+CYVL + +K +A+S K +
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMF 525
Query: 995 LGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLDSDQSKLVEKFADLS------ISVSDKG 1048
+GYS KG++ Y+ +A+ + S V+F ++ + K E DL+ + V +G
Sbjct: 524 IGYSTTQKGYKCYDPEARRVLVSRDVKFIEERGYYEEKNQEDLRDLTSDKAGVLRVILEG 345
Query: 1049 KALEEAEPEEDDPEEAGPSDSQPRKKSR 1076
++ + + + S ++PR+ ++
Sbjct: 344 LGIKMNQDQSTRSRQPEESSNEPRRAAQ 261
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 67.8 bits (164), Expect(2) = 1e-14
Identities = 41/125 (32%), Positives = 65/125 (51%), Gaps = 3/125 (2%)
Frame = +1
Query: 1270 QAPRAWYERLSSFLLENEFVRGKVDTTLFCK---TYKDDILIVQIYVDKIIFGSANQSLC 1326
Q+PR W++R + + + +++ + D +F K T K ILIV YVD I +
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIV--YVDDIFLTGDHGK*I 174
Query: 1327 KEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNMTESTIAKTP 1386
K ++ EFE+ +G LKYFLG++V + +G+ I Q KY +LLK+ M + P
Sbjct: 175 KRLKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCKTIRDP 354
Query: 1387 MHPTC 1391
C
Sbjct: 355 YGCNC 369
Score = 32.0 bits (71), Expect(2) = 1e-14
Identities = 18/52 (34%), Positives = 29/52 (55%)
Frame = +2
Query: 1383 AKTPMHPTCILEKEDASGKVCQKLYRCMIGSLLYLTASRPDNLFSVHLCARF 1434
++TPM T L D V + Y+ ++G L+YL+ +RPD F V ++F
Sbjct: 344 SETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQF 499
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 71.6 bits (174), Expect = 2e-12
Identities = 37/74 (50%), Positives = 48/74 (64%)
Frame = -2
Query: 1200 FVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPPGFEDEKNPDHVF 1259
FVP+ +L I L+S N+ L +DVK+AFL G + E++Y+HQP GF E V
Sbjct: 554 FVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGFS*EVG-KMVG 378
Query: 1260 KLKKSLYGLKQAPR 1273
KLKKS+YGLKQ PR
Sbjct: 377 KLKKSMYGLKQGPR 336
>BI262818 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (24%)
Length = 615
Score = 68.9 bits (167), Expect = 1e-11
Identities = 42/149 (28%), Positives = 84/149 (56%), Gaps = 1/149 (0%)
Frame = +1
Query: 9 YNAKP-PMFDGQRFEYWKDRLESFFLGFDADLWDIIVDGYERPVDAEGKKIPRSEMTAEQ 67
Y A P P+FDG+ + W R+E++ DLW+ + + YE ++ + + + E+
Sbjct: 160 YKAPPLPVFDGENYHIWAARIEAYLEA--NDLWEAVEEDYEVLPLSDNPTMAQIKNHKER 333
Query: 68 KKLYAQHHKARAILLSAISYEEYQKITDLEFANGIFESLKMSHEGNKKVKESKTLSLIQK 127
K + KARA L +A+S E + +I ++ A I+ LK +EG+++++ + L+LI++
Sbjct: 334 K---TRKSKARATLFAAVSEEIFTRIMTIKSAFEIWNFLKTEYEGDERIRGMQALNLIRE 504
Query: 128 YESFIMEPNESIEEMFSRFQLLVAGIRPL 156
+E M+ +E+I+E ++ + +R L
Sbjct: 505 FEMQKMKESETIKEYANKLISIANKVRLL 591
>BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T15F17.l
{Arabidopsis thaliana}, partial (3%)
Length = 539
Score = 63.2 bits (152), Expect = 8e-10
Identities = 36/104 (34%), Positives = 53/104 (50%)
Frame = -3
Query: 1415 LYLTASRPDNLFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGY 1474
+ LT P+ FS++L +R+ S P H +K I +YLKG ++GL Y K L GY
Sbjct: 531 ILLTLQGPNITFSINLLSRYSSAPTMRH*NGIKHICKYLKGIIDMGLFYSKDCSPDLIGY 352
Query: 1475 CDVDYAGDRT*RKSTFGNCQFLGSNLVSWASKRQSTIELSTAKA 1518
+ Y D +S G G+ ++SW S + STI S+ A
Sbjct: 351 VNA*YLSDPHKARS*TGYIFTCGNTVISWRSTK*STIATSSNHA 220
>BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse transcriptase
homolog - rape retrotransposon copia-like (fragment),
partial (84%)
Length = 249
Score = 58.5 bits (140), Expect = 2e-08
Identities = 25/46 (54%), Positives = 33/46 (71%)
Frame = -1
Query: 1237 ISEEVYVHQPPGFEDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSF 1282
+ E++Y+ QP GF DHV KL+KSLYGLKQ+PR WY+R S+
Sbjct: 246 LEEKIYMTQPEGFLFPGKEDHVCKLRKSLYGLKQSPRQWYKRFDSY 109
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.136 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,719,169
Number of Sequences: 36976
Number of extensions: 693538
Number of successful extensions: 4080
Number of sequences better than 10.0: 129
Number of HSP's better than 10.0 without gapping: 3864
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4037
length of query: 1622
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1513
effective length of database: 4,984,343
effective search space: 7541310959
effective search space used: 7541310959
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0346.14


