

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
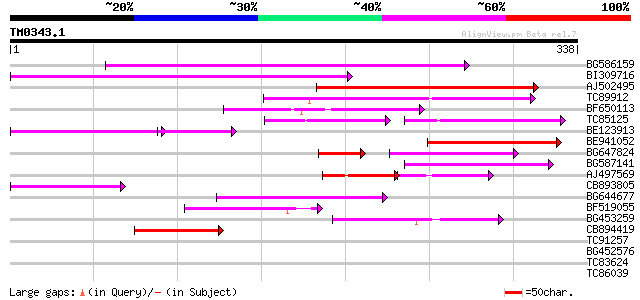
Query= TM0343.1
(338 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 160 8e-40
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 147 4e-36
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 127 5e-30
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 105 2e-23
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 100 6e-22
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 65 2e-21
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 62 2e-16
BE941052 weakly similar to PIR|B85188|B85 retrotransposon like p... 77 1e-14
BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F... 55 5e-13
BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440... 69 3e-12
AJ497569 weakly similar to PIR|T04833|T04 hypothetical protein F... 43 5e-10
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 55 3e-08
BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T... 53 1e-07
BF519055 similar to GP|21592754|gb| unknown {Arabidopsis thalian... 50 2e-06
BG453259 homologue to GP|21434|emb|CA ORF4 {Solanum tuberosum}, ... 49 2e-06
CB894419 weakly similar to GP|10177935|db copia-type polyprotein... 44 1e-04
TC91257 similar to GP|21434|emb|CAA36616.1|| ORF4 {Solanum tuber... 40 0.002
BG452576 weakly similar to GP|12005223|gb|A reverse transcriptas... 37 0.011
TC83624 homologue to PIR|G84581|G84581 copia-like retroelement p... 36 0.024
TC86039 similar to PIR|S66340|S66340 phloem-specific protein Vei... 35 0.041
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 160 bits (404), Expect = 8e-40
Identities = 82/217 (37%), Positives = 124/217 (56%)
Frame = +1
Query: 58 IAVTRHAGGLFLSQSTYASEIIARAGMASCNPSATPVDTKQKLSSSSGTPCEDATLYRSL 117
+ V ++ G+++ Q Y ++++ R GM N S P+ + KL DAT Y+ +
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 118 AGALQYLTFTRPDISYAVQQVCLHMHAPHTEHMLALKRVLRYVRGTLTYGLHLYPSPIAK 177
G L YL TRPD+ Y + + M+ P HM A+KRVLRY+ GT+ G+ + K
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 178 LVSYTDADWGGCPDTRRSTSGYCVFLGDNLISWSSKRQPTLSRSSAEAEYRGVANVVSES 237
L +YTD+D+ G D R+STSGY L +SWSSK+QP ++ S+ +AE+ A +S
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQS 540
Query: 238 CWIRNLLLELHFPLSQATLVHCDNVSAIYLSGNPVHH 274
W+R +L +L + S + ++CDN S I LS NPV H
Sbjct: 541 VWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 147 bits (372), Expect = 4e-36
Identities = 82/204 (40%), Positives = 111/204 (54%)
Frame = +2
Query: 1 DHSLFIFRRGTDIAYILLYVDDIILVASSHALRKSFMALLASEFAMKDLGPLSYFLGIAV 60
D SLF + + +L+YVDDI+L + + + L F +KDLG L YFLG+ V
Sbjct: 128 DFSLFTKFKDSSFTTLLVYVDDIVLAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEV 307
Query: 61 TRHAGGLFLSQSTYASEIIARAGMASCNPSATPVDTKQKLSSSSGTPCEDATLYRSLAGA 120
R G+ L+Q Y E++ +G + + TP D KL +S D T YR L G
Sbjct: 308 ARSKQGILLNQRKYTLELLEDSGNLAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGK 487
Query: 121 LQYLTFTRPDISYAVQQVCLHMHAPHTEHMLALKRVLRYVRGTLTYGLHLYPSPIAKLVS 180
L YLT TRPDIS+AVQQ+ + P H A RVL+Y++ GL + KL S
Sbjct: 488 LIYLTTTRPDISFAVQQLSQFVSKPQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSS 667
Query: 181 YTDADWGGCPDTRRSTSGYCVFLG 204
+ D+DW CP TR+S +GY VFLG
Sbjct: 668 FADSDWATCPTTRKSVTGYWVFLG 739
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 127 bits (320), Expect = 5e-30
Identities = 60/132 (45%), Positives = 87/132 (65%)
Frame = +2
Query: 184 ADWGGCPDTRRSTSGYCVFLGDNLISWSSKRQPTLSRSSAEAEYRGVANVVSESCWIRNL 243
+DW G +TR+STSGY LG ISWSSK+QP ++ S+AEAEY + +++ W+R +
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 244 LLELHFPLSQATLVHCDNVSAIYLSGNPVHHQRTKHIEMDIHFVREKVARGQARVLHVPS 303
L +H + T ++CDN SAI LS NPV H R+KHI++ H +RE +A + + + P+
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 304 RHQIADIFTKGL 315
+IADIFTK L
Sbjct: 362 EEKIADIFTKPL 397
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 105 bits (262), Expect = 2e-23
Identities = 57/164 (34%), Positives = 94/164 (56%), Gaps = 2/164 (1%)
Frame = +1
Query: 152 ALKRVLRYVRGTLTYGLHLYPSPIAK--LVSYTDADWGGCPDTRRSTSGYCVFLGDNLIS 209
ALK VL+Y+ +L L + + L Y DAD+ G DTR+S SG+ L IS
Sbjct: 4 ALKWVLKYLNESLKSSLKYTKAAQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTTIS 183
Query: 210 WSSKRQPTLSRSSAEAEYRGVANVVSESCWIRNLLLELHFPLSQATLVHCDNVSAIYLSG 269
W + +Q ++ S+ +AEY V ++ W++ ++ EL + +HCD+ SAI+L+
Sbjct: 184 WKANQQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGIT-QEYVKIHCDSQSAIHLAN 360
Query: 270 NPVHHQRTKHIEMDIHFVREKVARGQARVLHVPSRHQIADIFTK 313
+ V+H+RTKHI++ +HF+R+ + + V + S AD+FTK
Sbjct: 361 HQVYHERTKHIDIRLHFIRDMIESKEIVVEKMASEENPADVFTK 492
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 100 bits (250), Expect = 6e-22
Identities = 54/124 (43%), Positives = 75/124 (59%), Gaps = 4/124 (3%)
Frame = +1
Query: 128 RPDISYAVQQVCLHMHAPHTEHMLALKRVLRYVRGTLTYGLHLYP----SPIAKLVSYTD 183
RPDI Y+V + MH P H++A R+LRYVRGT+ YGL L+P S + +L+ Y+D
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGL-LFPYGAKSEVYELICYSD 297
Query: 184 ADWGGCPDTRRSTSGYCVFLGDNLISWSSKRQPTLSRSSAEAEYRGVANVVSESCWIRNL 243
+DW G RRSTSGY D ISW +K+QP + SS EAEY ++ W+ ++
Sbjct: 298 SDWCG---DRRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSV 468
Query: 244 LLEL 247
+ EL
Sbjct: 469 IKEL 480
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 65.1 bits (157), Expect(2) = 2e-21
Identities = 36/96 (37%), Positives = 53/96 (54%)
Frame = +3
Query: 236 ESCWIRNLLLELHFPLSQATLVHCDNVSAIYLSGNPVHHQRTKHIEMDIHFVREKVARGQ 295
E+ W++ L+ EL Q T V+CD+ SA++++ NP H RTKHI + HFVRE V G
Sbjct: 249 EAIWMQRLMEELGHKQEQIT-VYCDSQSALHIARNPAFHSRTKHIGIQYHFVREVVEEGS 425
Query: 296 ARVLHVPSRHQIADIFTKGLPRVLFDDFRSSLSVGE 331
+ + + +AD TK + F RSS + E
Sbjct: 426 VDMQKIHTNDNLADAMTKSINTDKFIWCRSSYGLLE 533
Score = 55.1 bits (131), Expect(2) = 2e-21
Identities = 28/75 (37%), Positives = 44/75 (58%)
Frame = +1
Query: 153 LKRVLRYVRGTLTYGLHLYPSPIAKLVSYTDADWGGCPDTRRSTSGYCVFLGDNLISWSS 212
+KR++RY++GT + S + + Y D+D+ G D R+ST+GY L +SW S
Sbjct: 1 VKRIMRYIKGTSGVAVCFGGSELT-VRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLS 177
Query: 213 KRQPTLSRSSAEAEY 227
K Q ++ S+ EAEY
Sbjct: 178 KLQTVVALSTTEAEY 222
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 61.6 bits (148), Expect(2) = 2e-16
Identities = 36/94 (38%), Positives = 47/94 (49%), Gaps = 1/94 (1%)
Frame = +1
Query: 1 DHSLFIFRRGT-DIAYILLYVDDIILVASSHALRKSFMALLASEFAMKDLGPLSYFLGIA 59
DH++FI T A +++YVDDI L K LLA EF +KDLG L YFLG+
Sbjct: 73 DHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKRLKNLLAEEFEIKDLGNLKYFLGME 252
Query: 60 VTRHAGGLFLSQSTYASEIIARAGMASCNPSATP 93
V R G +SQ Y +++ M C P
Sbjct: 253 VARWKKGSSISQRKYVLDLLKETRMIGCKTIRDP 354
Score = 41.2 bits (95), Expect(2) = 2e-16
Identities = 21/47 (44%), Positives = 26/47 (54%)
Frame = +2
Query: 89 PSATPVDTKQKLSSSSGTPCEDATLYRSLAGALQYLTFTRPDISYAV 135
PS TP+D KL + D Y+ L G L YL+ TRPDIS+ V
Sbjct: 341 PSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVV 481
>BE941052 weakly similar to PIR|B85188|B85 retrotransposon like protein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 480
Score = 76.6 bits (187), Expect = 1e-14
Identities = 37/80 (46%), Positives = 53/80 (66%)
Frame = +2
Query: 250 PLSQATLVHCDNVSAIYLSGNPVHHQRTKHIEMDIHFVREKVARGQARVLHVPSRHQIAD 309
P+SQ+ L+ CD +SA YL+ NPV+H R KHI +DIHFVR+ V +G+ +V HV + Q+AD
Sbjct: 2 PISQSGLLRCDYLSATYLTHNPVYHSRMKHISIDIHFVRDLVQQGKLKVQHVCTVDQLAD 181
Query: 310 IFTKGLPRVLFDDFRSSLSV 329
TK L + R+ + V
Sbjct: 182 CLTKPLSKSRHQLLRNKIGV 241
>BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (5%)
Length = 721
Score = 55.5 bits (132), Expect(2) = 5e-13
Identities = 28/78 (35%), Positives = 45/78 (56%), Gaps = 1/78 (1%)
Frame = -3
Query: 227 YRGVANVVSESCWIRNLLLELHFPLSQATLVHCDNVSAI-YLSGNPVHHQRTKHIEMDIH 285
YR + + + E W+ LL +L F + +++CDN SA +++ N +RTKHIE+D H
Sbjct: 374 YRSI*STICEIKWLTYLLNDLKFTFIKPAMLYCDNQSAARHIAANSSFLERTKHIELDCH 195
Query: 286 FVREKVARGQARVLHVPS 303
VR K+ +LH+ S
Sbjct: 194 IVRVKLQLKLFHILHILS 141
Score = 35.8 bits (81), Expect(2) = 5e-13
Identities = 15/28 (53%), Positives = 19/28 (67%)
Frame = -2
Query: 185 DWGGCPDTRRSTSGYCVFLGDNLISWSS 212
D C DT +S S +C+FLGD+LI W S
Sbjct: 486 D*SSCLDT*KSISYFCIFLGDSLICWKS 403
>BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440.1
[imported] - Arabidopsis thaliana, partial (20%)
Length = 731
Score = 68.9 bits (167), Expect = 3e-12
Identities = 35/89 (39%), Positives = 50/89 (55%)
Frame = +3
Query: 236 ESCWIRNLLLELHFPLSQATLVHCDNVSAIYLSGNPVHHQRTKHIEMDIHFVREKVARGQ 295
++ W+++LL E+ + + ++ DN S I L+ NPV H R HI HF+RE V GQ
Sbjct: 135 QAMWLQDLLSEVTWEPCEEVVIRIDNQSVIALTRNPVFHGRGNHIHKRYHFIRECVENGQ 314
Query: 296 ARVLHVPSRHQIADIFTKGLPRVLFDDFR 324
V HVP A I TK L R++F + R
Sbjct: 315 VEVEHVPGEKHRAYI*TKALGRIIFREIR 401
>AJ497569 weakly similar to PIR|T04833|T04 hypothetical protein F21P8.50 -
Arabidopsis thaliana, partial (4%)
Length = 723
Score = 42.7 bits (99), Expect(2) = 5e-10
Identities = 23/46 (50%), Positives = 31/46 (67%)
Frame = +3
Query: 187 GGCPDTRRSTSGYCVFLGDNLISWSSKRQPTLSRSSAEAEYRGVAN 232
G CP + R T+ +C FL +LISW SK+Q +SRS +EA R +AN
Sbjct: 84 GSCPYSIR*TTEFC-FLSSSLISWKSKKQCVVSRSFSEA**RALAN 218
Score = 38.5 bits (88), Expect(2) = 5e-10
Identities = 20/57 (35%), Positives = 33/57 (57%)
Frame = +2
Query: 232 NVVSESCWIRNLLLELHFPLSQATLVHCDNVSAIYLSGNPVHHQRTKHIEMDIHFVR 288
N+ C+I ++ H S L +CDN+SA++++ N V H+RT H E D + V+
Sbjct: 221 NLRLHGCFIFYMISAKH---ST*VLQYCDNISALHIAANMVFHERT*HRETDPYIVQ 382
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 55.5 bits (132), Expect = 3e-08
Identities = 26/70 (37%), Positives = 42/70 (59%), Gaps = 1/70 (1%)
Frame = +3
Query: 1 DHSLFI-FRRGTDIAYILLYVDDIILVASSHALRKSFMALLASEFAMKDLGPLSYFLGIA 59
+H+LF+ G I I LYVDD+I + + + + F + EF M DLG + YFLG+
Sbjct: 564 EHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGKMHYFLGVE 743
Query: 60 VTRHAGGLFL 69
VT++ G+++
Sbjct: 744 VTQNEKGIYI 773
>BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T15F17.l
{Arabidopsis thaliana}, partial (3%)
Length = 539
Score = 53.1 bits (126), Expect = 1e-07
Identities = 29/102 (28%), Positives = 51/102 (49%)
Frame = -3
Query: 124 LTFTRPDISYAVQQVCLHMHAPHTEHMLALKRVLRYVRGTLTYGLHLYPSPIAKLVSYTD 183
LT P+I++++ + + AP H +K + +Y++G + GL L+ Y +
Sbjct: 525 LTLQGPNITFSINLLSRYSSAPTMRH*NGIKHICKYLKGIIDMGLFYSKDCSPDLIGYVN 346
Query: 184 ADWGGCPDTRRSTSGYCVFLGDNLISWSSKRQPTLSRSSAEA 225
A + P RS +GY G+ +ISW S + T++ SS A
Sbjct: 345 A*YLSDPHKARS*TGYIFTCGNTVISWRSTK*STIATSSNHA 220
>BF519055 similar to GP|21592754|gb| unknown {Arabidopsis thaliana}, partial
(30%)
Length = 675
Score = 49.7 bits (117), Expect = 2e-06
Identities = 27/84 (32%), Positives = 47/84 (55%), Gaps = 2/84 (2%)
Frame = -3
Query: 105 GTPCEDATLYRSLAGALQYLTFTRPDISYAVQQVCLHMHAPHTEHMLALKRVLRYVRGTL 164
G+ D+ L S+ GAL Y+T T PD+S+++ + MH P + LK+V+R+ + T+
Sbjct: 661 GSITADSKLVHSIVGALLYITVTCPDLSFSINKPSQFMHKPTQINFQQLKKVMRHPKLTI 482
Query: 165 --TYGLHLYPSPIAKLVSYTDADW 186
+ L +Y +++DADW
Sbjct: 481 KKLFDLQIY--------AFSDADW 434
>BG453259 homologue to GP|21434|emb|CA ORF4 {Solanum tuberosum}, partial (5%)
Length = 657
Score = 49.3 bits (116), Expect = 2e-06
Identities = 35/105 (33%), Positives = 56/105 (53%), Gaps = 3/105 (2%)
Frame = -2
Query: 193 RRSTSGYCVFLGDNLISWSSKRQPT-LSRSSAEAEYRGVANVVSESCWIR--NLLLELHF 249
R STSGY +FLG N++ + R + E +G+ V+ E I+ +L++
Sbjct: 455 RGSTSGY*MFLGGNMVE*KQNVVAR*VQRHNFELCSQGL*RVMDEELKIKLDDLIINYKD 276
Query: 250 PLSQATLVHCDNVSAIYLSGNPVHHQRTKHIEMDIHFVREKVARG 294
P++ + +N ++ NPV H RTKHIE+D HF+ EK+ G
Sbjct: 275 PMT----LF*NNNFVSRIAHNPVQHYRTKHIEIDQHFIIEKLYSG 153
>CB894419 weakly similar to GP|10177935|db copia-type polyprotein
{Arabidopsis thaliana}, partial (2%)
Length = 170
Score = 43.5 bits (101), Expect = 1e-04
Identities = 23/53 (43%), Positives = 33/53 (61%)
Frame = +3
Query: 75 ASEIIARAGMASCNPSATPVDTKQKLSSSSGTPCEDATLYRSLAGALQYLTFT 127
AS+I+ + M P +TPV+ K KL+ S D+T Y+SL G+L+YLT T
Sbjct: 12 ASDILKKFKMEHSKPISTPVEEKLKLTRESDGKRVDSTHYKSLIGSLRYLTAT 170
>TC91257 similar to GP|21434|emb|CAA36616.1|| ORF4 {Solanum tuberosum},
partial (8%)
Length = 854
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/34 (58%), Positives = 20/34 (58%)
Frame = +3
Query: 105 GTPCEDATLYRSLAGALQYLTFTRPDISYAVQQV 138
G D YR L G L YLT TRPDISYAV V
Sbjct: 735 GETFSDPGRYRRLVGKLNYLTMTRPDISYAVSVV 836
>BG452576 weakly similar to GP|12005223|gb|A reverse transcriptase-like
protein {Spiranthes hongkongensis}, partial (25%)
Length = 676
Score = 37.0 bits (84), Expect = 0.011
Identities = 24/83 (28%), Positives = 39/83 (46%)
Frame = +2
Query: 1 DHSLFIFRRGTDIAYILLYVDDIILVASSHALRKSFMALLASEFAMKDLGPLSYFLGIAV 60
DHS+F F R I L+Y+DDI+ H+ + + L F + LG L Y +G+ +
Sbjct: 299 DHSIFSFGRRFTI--FLVYLDDIVFARDDHSETQLVKSHLDKNFIIIGLGTLHY-VGVKI 469
Query: 61 TRHAGGLFLSQSTYASEIIARAG 83
+ Q Y E++ +G
Sbjct: 470 A*SES*IIDDQCKYTIELLEESG 538
>TC83624 homologue to PIR|G84581|G84581 copia-like retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(1%)
Length = 831
Score = 35.8 bits (81), Expect = 0.024
Identities = 20/55 (36%), Positives = 32/55 (57%), Gaps = 1/55 (1%)
Frame = +2
Query: 244 LLELHFPLSQATLVHCDNVSAIYLSGNPVHHQRTKHIEMDIHFVR-EKVARGQAR 297
L L ++ L++C N +Y++ N V+H+RTKH E + F+ EKVA +R
Sbjct: 443 LRNLQVQCTRLLLIYCVNQITLYIAKNQVYHERTKH*ENNWTFLF*EKVANWGSR 607
>TC86039 similar to PIR|S66340|S66340 phloem-specific protein Vein1 - fava
bean, partial (59%)
Length = 1180
Score = 35.0 bits (79), Expect = 0.041
Identities = 16/29 (55%), Positives = 20/29 (68%)
Frame = +2
Query: 278 KHIEMDIHFVREKVARGQARVLHVPSRHQ 306
KHIE DIHFV E+V Q +LHVP ++
Sbjct: 710 KHIEFDIHFV*ERVIAKQLVMLHVPRSYK 796
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.135 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,256,596
Number of Sequences: 36976
Number of extensions: 169531
Number of successful extensions: 860
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 848
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 854
length of query: 338
length of database: 9,014,727
effective HSP length: 97
effective length of query: 241
effective length of database: 5,428,055
effective search space: 1308161255
effective search space used: 1308161255
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0343.1


