

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0342a.5
(1069 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
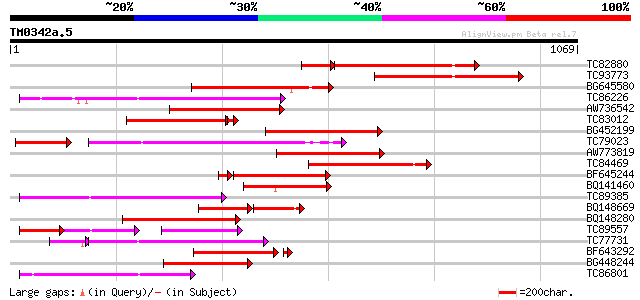
Score E
Sequences producing significant alignments: (bits) Value
TC82880 similar to GP|9858483|gb|AAG01054.1| resistance protein ... 322 e-103
TC93773 weakly similar to GP|9858483|gb|AAG01054.1| resistance p... 330 2e-90
BG645580 similar to PIR|T07656|T076 probable resistance protein ... 321 1e-87
TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 313 3e-85
AW736542 similar to PIR|T07656|T076 probable resistance protein ... 297 1e-80
TC83012 weakly similar to GP|7107242|gb|AAF36335.1| unknown {Cic... 253 7e-70
BG452199 weakly similar to GP|10178211|db TMV resistance protein... 261 1e-69
TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 256 4e-68
AW773819 similar to GP|9858483|gb|A resistance protein MG55 {Gly... 249 4e-66
TC84469 similar to GP|9858483|gb|AAG01054.1| resistance protein ... 247 2e-65
BF645244 weakly similar to PIR|T07656|T076 probable resistance p... 224 1e-64
BQ141460 similar to PIR|T07656|T076 probable resistance protein ... 240 2e-63
TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resist... 239 3e-63
BQ148669 similar to PIR|T07656|T076 probable resistance protein ... 138 8e-62
BQ148280 similar to GP|7107246|gb| unknown {Cicer arietinum}, pa... 228 7e-60
TC89557 weakly similar to GP|10178211|dbj|BAB11635. TMV resistan... 102 1e-58
TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 221 1e-57
BF643292 weakly similar to GP|22037340|gb| disease resistance-li... 214 7e-56
BG448244 weakly similar to PIR|T08833|T088 disease resistance pr... 213 2e-55
TC86801 similar to PIR|A54810|A54810 TMV resistance protein N - ... 208 8e-54
>TC82880 similar to GP|9858483|gb|AAG01054.1| resistance protein MG55
{Glycine max}, partial (46%)
Length = 1238
Score = 322 bits (825), Expect(2) = e-103
Identities = 172/275 (62%), Positives = 211/275 (76%), Gaps = 2/275 (0%)
Frame = +2
Query: 613 LLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIGDLSNLLLINLEGC 672
L+E LKIL+LSHS YL+ TPDFSKLPNLEKLI+KDC SL E+H +IGDL+NLLLINL+ C
Sbjct: 296 LIEGLKILNLSHSKYLKRTPDFSKLPNLEKLIMKDCQSLLEVHPSIGDLNNLLLINLKDC 475
Query: 673 TSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIAKDTAIKEVPYSILGL 732
TSL+NLP + YQL++VKTLI+SGCSKIDKLEEDI QMESLTTL+A +T +K+ P+SI+
Sbjct: 476 TSLSNLPREIYQLRTVKTLILSGCSKIDKLEEDIVQMESLTTLMAANTGVKQPPFSIVRS 655
Query: 733 KSIGYISLCGYEGLSRDVFPSLLRSWISPTMNPLSHIPLFAAMSLPLVSMDIQ--NYSSG 790
KSIGYISLCGYEGLS VFPSL+RSW+SPTMN ++HI F MS L S+DI+ N +
Sbjct: 656 KSIGYISLCGYEGLSHHVFPSLIRSWMSPTMNSVAHISPFGGMSKSLASLDIESNNLALV 835
Query: 791 NSSSWLSSLSKLRSVWIQCHSEIQLAQESRRILDDQYDVNSMDLEASSSSSYASQISDFS 850
S LSS SKLRSV +QC SEIQL QE RR LDD YD +L S+AS ISD S
Sbjct: 836 YQSQILSSCSKLRSVSVQCDSEIQLKQEFRRFLDDLYDAGLTEL----GISHASHISDHS 1003
Query: 851 LKSLLIRLGSCYTVIDTLASSISQGSTTNASSNFF 885
L+SLLI +G+C+ VI+ L S+SQ + ++ F+
Sbjct: 1004LRSLLIGMGNCHIVINILGKSLSQVPSQSSHLIFY 1108
Score = 72.4 bits (176), Expect(2) = e-103
Identities = 30/64 (46%), Positives = 48/64 (74%)
Frame = +3
Query: 550 KLRLLQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYSSVKQVWK 609
KLRLLQL+ V + GDY S +L+W+ W+GF Y+P++FY N+V ++LK+S++ QVWK
Sbjct: 3 KLRLLQLDNVQVIGDYECFSKQLRWLSWQGFPLKYMPENFYQKNVVAMDLKHSNLTQVWK 182
Query: 610 ETKL 613
+ ++
Sbjct: 183 KPQV 194
>TC93773 weakly similar to GP|9858483|gb|AAG01054.1| resistance protein MG55
{Glycine max}, partial (31%)
Length = 856
Score = 330 bits (845), Expect = 2e-90
Identities = 170/282 (60%), Positives = 213/282 (75%), Gaps = 2/282 (0%)
Frame = +2
Query: 689 KTLIISGCSKIDKLEEDIAQMESLTTLIAKDTAIKEVPYSILGLKSIGYISLCGYEGLSR 748
+ I+SGCSKI+KLEEDI QM+SLTTLIA T +K+VP+SI+ K+IGYISLC YEGLSR
Sbjct: 17 ENFILSGCSKIEKLEEDIVQMKSLTTLIAAKTGVKQVPFSIVKSKNIGYISLCEYEGLSR 196
Query: 749 DVFPSLLRSWISPTMNPLSHIPLFAAMSLPLVSMDIQNYSSG--NSSSWLSSLSKLRSVW 806
DVFPS++ SW+SP MN L+HIP MS+ LV +D+ + + G + S LSS SKLRSV
Sbjct: 197 DVFPSIIWSWMSPNMNSLAHIPPVGGMSMSLVCLDVDSRNLGLVHQSPILSSYSKLRSVS 376
Query: 807 IQCHSEIQLAQESRRILDDQYDVNSMDLEASSSSSYASQISDFSLKSLLIRLGSCYTVID 866
+QC SEIQL QE RR LDD YD + +S+ SQI D SL+SLL +GSC+ VI+
Sbjct: 377 VQCDSEIQLKQEFRRFLDDIYDAGLTEF----GTSHGSQILDLSLRSLLFGIGSCHIVIN 544
Query: 867 TLASSISQGSTTNASSNFFLPGGDDPYWLAYKGEGPSVHFQVPEDRDCCLKGIVLCVVYS 926
TL S+S+G N SS FLPG + P WLAY+GEGPS F+VPED DC +KG+ LCV+YS
Sbjct: 545 TLRKSLSEGLAAN-SSGSFLPGDNYPSWLAYRGEGPSAIFKVPEDTDCRMKGMTLCVLYS 721
Query: 927 STPENMATECLTSVMINNHTKFTIQIYKQGTVMSFNDEDWES 968
ST +N+ATECLT V+I N+TKFTIQIYK+ TVMSFNDE+W++
Sbjct: 722 STSKNVATECLTGVLIINYTKFTIQIYKRHTVMSFNDEEWQA 847
>BG645580 similar to PIR|T07656|T076 probable resistance protein - soybean
(fragment), partial (87%)
Length = 814
Score = 321 bits (822), Expect = 1e-87
Identities = 168/274 (61%), Positives = 202/274 (73%), Gaps = 7/274 (2%)
Frame = +1
Query: 344 ESLELFSWHAFGEASPREDFNELSRRVVAYCGGLPLALEVLGSYLNEREEIEWESVLSKL 403
+ LELFSWHAF + SP ++F+ELSR VVAYCGGLPLALEV+GSYL R + EWESVL KL
Sbjct: 1 DPLELFSWHAFRQPSPIKNFSELSRTVVAYCGGLPLALEVIGSYLYGRTKQEWESVLLKL 180
Query: 404 ERIPNDQVQQKLRISYDGLKDELEKDIFLDICCFFVGKDRAYVTDILNGCGLYADIGITL 463
ERIPNDQVQ+KLRISYDGLKD++ KDIFLDICCFF+GKDRAYVT+ILNGCGLYADIGIT+
Sbjct: 181 ERIPNDQVQEKLRISYDGLKDDMAKDIFLDICCFFIGKDRAYVTEILNGCGLYADIGITV 360
Query: 464 LIERSLLQVEKNNKLGMHDLVRDMGRAIVRESSTKDLGKRSRLWFHEDVHDVLTKNTGTE 523
L+ERSL+++EKNNKLGMHDL+RDMGR IVR+SS K+ GKRSRLWFHEDVHDVLTKNTGT
Sbjct: 361 LVERSLVKIEKNNKLGMHDLLRDMGREIVRQSSAKNPGKRSRLWFHEDVHDVLTKNTGTI 540
Query: 524 TVEGLIL-------KLQSTSRVCFSTNAFKEMRKLRLLQLECVDLTGDYRHLSHELKWVY 576
VEG++ + S + + N L+ L +++ G + +
Sbjct: 541 NVEGVVF*VCQEPAEFASVLILSWK*NN*NN*NFYNLIVLTSLEIMGVF---PNN*DGSV 711
Query: 577 WKGFTSTYIPDHFYLGNLVVLELKYSSVKQVWKE 610
K T F NLV L+LK+S +KQVW E
Sbjct: 712 CKDLP*TVYQMTFIRKNLVALDLKHSKIKQVWNE 813
>TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1769
Score = 313 bits (801), Expect = 3e-85
Identities = 194/519 (37%), Positives = 299/519 (57%), Gaps = 16/519 (3%)
Frame = +3
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEK--LKKGSELKPALLRAIEGSRISLAVLSSNY 75
G DTR+ F +L AL + GI F DD+ L++ ++ P + IE SRI + + S+NY
Sbjct: 114 GTDTRYGFTGNLLKALIDKGIRTFHDDDDSDLQRRDKVTPII---IEESRILIPIFSANY 284
Query: 76 ASSRWCLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQKGPVLLLGKQLQVE------D 129
ASS CLD LV I+ C +T G LV+P+F+ V+P+ VR+ G GK L D
Sbjct: 285 ASSSSCLDTLVHIIHCYKTKGCLVLPVFFGVEPTDVRHHTGRY---GKALAEHENRFQND 455
Query: 130 TQRKTRRGGHEVA-----SFSGYNLSPDRSEAEAINEIVKNVFRNLDIRLMSITDFPVGL 184
T+ R +VA + Y+ E E I +IVK + + + + + +PVGL
Sbjct: 456 TKNMERLQQWKVALSLAANLPSYHDDSHGYEYELIGKIVKYISNKISRQSLHVATYPVGL 635
Query: 185 ESHLQEVIKLI-ESQSSKVCMVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVC 243
+S +Q+V L+ E V MVGI+G+GG GK+T A+AI+N + +F F+E +RE
Sbjct: 636 QSRVQQVKSLLDEGPDDGVHMVGIYGIGGSGKSTLARAIYNFVADQFEGLCFLEQVRE-- 809
Query: 244 ENDSRGHMHLQEQLLSDVLKTKEKIHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRAL 303
+ S QE LLS L+ K K+ + G ++IK+ L K+ L+ILDDV +QL AL
Sbjct: 810 NSASNSLKRFQEMLLSKTLQLKIKLADVSEGISIIKERLCRKKILLILDDVDNMKQLNAL 989
Query: 304 CGNREWFASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPREDF 363
G +WF GS +I+TTRD LL + + Y +K ++ E+LEL W AF +
Sbjct: 990 AGGVDWFGPGSRVIITTRDKHLLACHEIEKTYAVKGLNVTEALELLRWMAFKNDKVPSSY 1169
Query: 364 NELSRRVVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGLK 423
++ RVVAY GLP+ +E++GS L + E ++ L E+IPN ++Q+ L++SYD L+
Sbjct: 1170EKILNRVVAYASGLPVVIEIVGSNLFGKNIEECKNTLDWYEKIPNKEIQRILKVSYDSLE 1349
Query: 424 DELEKDIFLDICCFFVGKDRAYVTDILNG-CGLYADIGITLLIERSLL-QVEKNNKLGMH 481
+E E+ +FLDI C F G V +IL+ G + + +L+E+ L+ E ++ + +H
Sbjct: 1350EE-EQSVFLDIACCFKGCKWEKVKEILHAHYGHCINHHVEVLVEKCLIDHFEYDSHVSLH 1526
Query: 482 DLVRDMGRAIVRESSTKDLGKRSRLWFHEDVHDVLTKNT 520
+L+ +MG+ +VR S + GKRSRLWF +D+ +VL +NT
Sbjct: 1527NLIENMGKELVRLESPFEPGKRSRLWFEKDIFEVLEENT 1643
>AW736542 similar to PIR|T07656|T076 probable resistance protein - soybean
(fragment), partial (83%)
Length = 656
Score = 297 bits (761), Expect = 1e-80
Identities = 144/217 (66%), Positives = 178/217 (81%)
Frame = +2
Query: 302 ALCGNREWFASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPRE 361
ALCGNR GS+II+TTRD RLL+ L D++Y + ++ ES LF+WHAF EA+P E
Sbjct: 5 ALCGNRNGIGPGSIIIITTRDARLLDILGVDFIYEAEGLNVHESRRLFNWHAFKEANPSE 184
Query: 362 DFNELSRRVVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDG 421
F LS VV+YCGGLPLALEVLGSYL R + EW+SV+SKL++IPNDQ+ +KL+IS+DG
Sbjct: 185 AFLILSGDVVSYCGGLPLALEVLGSYLFNRRKREWQSVISKLQKIPNDQIHEKLKISFDG 364
Query: 422 LKDELEKDIFLDICCFFVGKDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKLGMH 481
L+D +EK+IFLD+CCFF+GKDRAYVT+ILNGCGL+ADIGI +LIERSLL+VEKNNKLGMH
Sbjct: 365 LEDHMEKNIFLDVCCFFIGKDRAYVTEILNGCGLHADIGIEVLIERSLLKVEKNNKLGMH 544
Query: 482 DLVRDMGRAIVRESSTKDLGKRSRLWFHEDVHDVLTK 518
L+RDMGR IVRESS ++ KR+RLW EDV DVL +
Sbjct: 545 ALLRDMGREIVRESSPEEPEKRTRLWCFEDVVDVLAE 655
>TC83012 weakly similar to GP|7107242|gb|AAF36335.1| unknown {Cicer
arietinum}, partial (88%)
Length = 654
Score = 253 bits (647), Expect(2) = 7e-70
Identities = 126/198 (63%), Positives = 157/198 (78%), Gaps = 2/198 (1%)
Frame = +1
Query: 221 AIFNQIHRKFMCTSFIENIREVCENDSRG--HMHLQEQLLSDVLKTKEKIHSIGRGTAMI 278
AI+NQIHRKF+ SFIENIRE CE DS+G H+ LQ+QLLSD+LKTKEKIH+I GT I
Sbjct: 1 AIYNQIHRKFVYRSFIENIRETCERDSKGGWHICLQQQLLSDLLKTKEKIHNIASGTIAI 180
Query: 279 KKILSGKRALVILDDVTTSEQLRALCGNREWFASGSVIIVTTRDVRLLNSLKADYVYTMK 338
KK+LS K+ L++LDDVT EQ++AL +R+WF +GSV+IVT+RD +L SL+ D+VY +
Sbjct: 181 KKMLSAKKVLIVLDDVTKVEQVKALYESRKWFGAGSVLIVTSRDAHILKSLQVDHVYPVN 360
Query: 339 EMDEDESLELFSWHAFGEASPREDFNELSRRVVAYCGGLPLALEVLGSYLNEREEIEWES 398
EMD+ ESLELFSWHAF +ASPR DF+ELS V+ YCGGLPLA EV+GSYL R EW S
Sbjct: 361 EMDQKESLELFSWHAFRQASPRADFSELSSSVIKYCGGLPLAAEVIGSYLYGRTREEWTS 540
Query: 399 VLSKLERIPNDQVQQKLR 416
VLSKLE IP+ VQ++ +
Sbjct: 541 VLSKLEIIPDHHVQRETK 594
Score = 30.4 bits (67), Expect(2) = 7e-70
Identities = 13/19 (68%), Positives = 16/19 (83%)
Frame = +2
Query: 412 QQKLRISYDGLKDELEKDI 430
++KLRISYDGL D +KDI
Sbjct: 581 KEKLRISYDGLSDGKQKDI 637
>BG452199 weakly similar to GP|10178211|db TMV resistance protein N
{Arabidopsis thaliana}, partial (4%)
Length = 666
Score = 261 bits (667), Expect = 1e-69
Identities = 134/221 (60%), Positives = 162/221 (72%)
Frame = +3
Query: 483 LVRDMGRAIVRESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFST 542
L+RDMGR IVRE S K+ +RSRLWF++DV DVL++ TGT+ VEGL LKL + + CFST
Sbjct: 3 LLRDMGREIVREKSPKEPEERSRLWFNKDVIDVLSEKTGTKAVEGLALKLPTANGKCFST 182
Query: 543 NAFKEMRKLRLLQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYS 602
AFKEM+KLRLLQL V L GD+ +LS L W+ W GF IP FY NLV +EL S
Sbjct: 183 KAFKEMKKLRLLQLASVQLEGDFEYLSRNLIWLSWNGFPLKCIPSSFYQENLVSIELVNS 362
Query: 603 SVKQVWKETKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIGDLS 662
+VK VWKE + LEKLKIL+LSHS +L TPDFS LPNLE+L+L DC L E+ +IGDL+
Sbjct: 363 NVKVVWKEPQRLEKLKILNLSHSHFLSKTPDFSNLPNLEQLVLTDCPKLSEVSQSIGDLN 542
Query: 663 NLLLINLEGCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLE 703
+LLINLE C SL +LP Y LKS+ TLI+SGC IDKLE
Sbjct: 543 KILLINLEDCISLQSLPRSIYNLKSLNTLILSGCLMIDKLE 665
>TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (37%)
Length = 2072
Score = 256 bits (653), Expect = 4e-68
Identities = 170/491 (34%), Positives = 268/491 (53%), Gaps = 4/491 (0%)
Frame = +3
Query: 149 LSPDRSEAEAINEIVKNVFRNLDIRLMSITDFPVGLESHLQEVI-KLIESQSSKVCMVGI 207
L +R E E I +IV+++ + + + +PVGL+S +Q V L E +V MVG+
Sbjct: 675 LDMNRYEYEFIGKIVEDISNRISREPLDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMVGL 854
Query: 208 WGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCENDSRGHMHLQEQLLSDVLKTKEK 267
+G GG+GK+T AKAI+N I +F F+EN+R +D+ HLQE+LL ++ K
Sbjct: 855 YGTGGIGKSTLAKAIYNFIADQFEVLCFLENVRVNSTSDNL--KHLQEKLLLKTVRLDIK 1028
Query: 268 IHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALCGNREWFASGSVIIVTTRDVRLLN 327
+ + +G +IK+ L K+ L+ILDDV +QL AL G +WF GS +I+TTR+ LL
Sbjct: 1029 LGGVSQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKHLLK 1208
Query: 328 SLKADYVYTMKEMDEDESLELFSWHAFGEASPREDFNELSRRVVAYCGGLPLALEVLGSY 387
+ + ++ ++ E+LEL W AF E P ++ R + Y GLPLA+ ++GS
Sbjct: 1209 IHGIESTHAVEGLNATEALELLRWMAFKENVP-SSHEDILNRALTYASGLPLAIVIIGSN 1385
Query: 388 LNEREEIEWESVLSKLERIPNDQVQQKLRISYDGLKDELEKDIFLDICCFFVGKDRAYVT 447
L R + S L E IPN ++Q+ L++SYD L+ E E+ +FLDI C F G V
Sbjct: 1386 LVGRSVQDSMSTLDGYEEIPNKEIQRILKVSYDSLEKE-EQSVFLDIACCFKGCKWPEVK 1562
Query: 448 DILNG-CGLYADIGITLLIERSLL-QVEKNNKLGMHDLVRDMGRAIVRESSTKDLGKRSR 505
+IL+ G + +L E+SL+ ++ ++ + +HDL+ DMG+ +VR+ S + G+RSR
Sbjct: 1563 EILHAHYGHCIVHHVAVLAEKSLMDHLKYDSYVTLHDLIEDMGKEVVRQESPDEPGERSR 1742
Query: 506 LWFHEDVHDVLTKNTGTETVEGLILKLQS-TSRVCFSTNAFKEMRKLRLLQLECVDLTGD 564
LWF D+ VL KNTGT ++ + +K S S + ++ NAF++M L+ E
Sbjct: 1743 LWFERDIVHVLKKNTGTRKIKMINMKFPSMESDIDWNGNAFEKMTNLKTFITE------- 1901
Query: 565 YRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYSSVKQVWKETKLLEKLKILDLSH 624
H S L+ Y+P + + + SS K E +K+L L++
Sbjct: 1902 NGHHSKSLE----------YLPSSLRVMKGCIPKSPSSS-----SSNKKFEDMKVLILNN 2036
Query: 625 SWYLENTPDFS 635
YL + PD S
Sbjct: 2037 CEYLTHIPDVS 2069
Score = 107 bits (267), Expect = 2e-23
Identities = 54/106 (50%), Positives = 70/106 (65%)
Frame = +1
Query: 11 QLAMQAGGEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAV 70
Q+ + G DTRH F +LY AL++ GI F+DD L++G E+ P+L AIE SRI + V
Sbjct: 97 QVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKNAIEKSRIFIPV 276
Query: 71 LSSNYASSRWCLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQKG 116
S NYASS +CLDELV I C T G LV+P+F VDP+ VR+ G
Sbjct: 277 FSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHTG 414
>AW773819 similar to GP|9858483|gb|A resistance protein MG55 {Glycine max},
partial (20%)
Length = 633
Score = 249 bits (636), Expect = 4e-66
Identities = 124/204 (60%), Positives = 151/204 (73%)
Frame = +1
Query: 503 RSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFSTNAFKEMRKLRLLQLECVDLT 562
RSRLWF +DV DVL++ GT+ VEGL LKL + CFST AFK+M KLRLLQL V L
Sbjct: 19 RSRLWFDKDVFDVLSEQNGTKVVEGLALKLPRENAKCFSTKAFKKMEKLRLLQLAGVQLD 198
Query: 563 GDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYSSVKQVWKETKLLEKLKILDL 622
GD+ HLS L+W+ W GF T IP FY GNLV +EL S++K VWK+T+ LEKLKIL+L
Sbjct: 199 GDFEHLSRNLRWLSWNGFPLTCIPSSFYQGNLVSIELVNSNIKLVWKKTQRLEKLKILNL 378
Query: 623 SHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIGDLSNLLLINLEGCTSLNNLPGKT 682
SHS YL TPDFS LPNLE+L+L DC L E+ H+I L+ +LLINLE C L +LP
Sbjct: 379 SHSHYLTQTPDFSNLPNLEQLVLTDCPRLSEVSHSIEHLNKILLINLEDCIGLQSLPRSI 558
Query: 683 YQLKSVKTLIISGCSKIDKLEEDI 706
Y+LKS+KTLI+SGCS IDK+EED+
Sbjct: 559 YKLKSLKTLILSGCSMIDKVEEDL 630
>TC84469 similar to GP|9858483|gb|AAG01054.1| resistance protein MG55
{Glycine max}, partial (39%)
Length = 735
Score = 247 bits (630), Expect = 2e-65
Identities = 125/232 (53%), Positives = 167/232 (71%)
Frame = +3
Query: 563 GDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYSSVKQVWKETKLLEKLKILDL 622
GD++++S LKW++W GF IP +FY N+V +EL+ S+ K VWKE + +E+LKIL+L
Sbjct: 27 GDFKYISRNLKWLHWNGFPLRCIPSNFYQRNIVSIELENSNAKLVWKEIQRMEQLKILNL 206
Query: 623 SHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIGDLSNLLLINLEGCTSLNNLPGKT 682
SHS +L TPDFS LPNLEKL+L+DC L ++ H+IG L ++LINL+ C SL +LP
Sbjct: 207 SHSHHLTQTPDFSYLPNLEKLVLEDCPRLSQVSHSIGHLKKVVLINLKDCISLCSLPRNI 386
Query: 683 YQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIAKDTAIKEVPYSILGLKSIGYISLCG 742
Y LK++ TLI+SGC IDKLEED+ QMESLTTLIA +T I +VP+S++ KSIG+ISLCG
Sbjct: 387 YTLKTLNTLILSGCLMIDKLEEDLEQMESLTTLIANNTGITKVPFSLVRSKSIGFISLCG 566
Query: 743 YEGLSRDVFPSLLRSWISPTMNPLSHIPLFAAMSLPLVSMDIQNYSSGNSSS 794
YEG SRDVFPS++ SW+SP N LS A+ LVS++ + SS
Sbjct: 567 YEGFSRDVFPSIIWSWMSP--NNLSPAFQTASHMSSLVSLEASTLXXXDLSS 716
>BF645244 weakly similar to PIR|T07656|T076 probable resistance protein -
soybean (fragment), partial (81%)
Length = 678
Score = 224 bits (572), Expect(2) = 1e-64
Identities = 103/184 (55%), Positives = 144/184 (77%)
Frame = +3
Query: 422 LKDELEKDIFLDICCFFVGKDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKLGMH 481
L D+ EK+IFLD+ CFF+G DR VT ILNGC L+ +IGI++L+ERSL+ V+ NKLGMH
Sbjct: 87 LNDDFEKEIFLDVACFFIGMDRNDVTLILNGCELFGEIGISILVERSLVTVDGKNKLGMH 266
Query: 482 DLVRDMGRAIVRESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFS 541
DL+RDMGR I+RE S +++ +R RLWFH+DV VL++ TGT+T++GL LKLQ + CFS
Sbjct: 267 DLLRDMGREIIREKSPEEIEERCRLWFHDDVLHVLSEQTGTKTIKGLALKLQRANEKCFS 446
Query: 542 TNAFKEMRKLRLLQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKY 601
T AFK+M++LRLLQL V L GD++++S L+W+ W F+ST+IP +FY+ NL +EL+
Sbjct: 447 TKAFKKMKRLRLLQLAGVKLVGDFKYISRSLRWLSWNRFSSTHIPTNFYIXNLXSIELEN 626
Query: 602 SSVK 605
S++K
Sbjct: 627 SNIK 638
Score = 42.0 bits (97), Expect(2) = 1e-64
Identities = 18/25 (72%), Positives = 22/25 (88%)
Frame = +1
Query: 395 EWESVLSKLERIPNDQVQQKLRISY 419
EW+S L KL+RIPN+QV +KLRISY
Sbjct: 4 EWKSALDKLKRIPNNQVHKKLRISY 78
>BQ141460 similar to PIR|T07656|T076 probable resistance protein - soybean
(fragment), partial (68%)
Length = 523
Score = 240 bits (612), Expect = 2e-63
Identities = 117/174 (67%), Positives = 144/174 (82%), Gaps = 8/174 (4%)
Frame = +2
Query: 441 KDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKLGMHDLVRDMGRAIVRESSTKDL 500
KDRAYVT+ILNGCGL+A IGI++LIERSLL+VEKNNKLGMHDL+RDMGR IVR++S KD+
Sbjct: 2 KDRAYVTEILNGCGLFASIGISVLIERSLLKVEKNNKLGMHDLIRDMGREIVRQNSEKDV 181
Query: 501 --------GKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFSTNAFKEMRKLR 552
G+RSRLWF +DVHDVLT NTGT+TVEGL+L L++TSR F+T+AF EM+KLR
Sbjct: 182 RQISEKDPGERSRLWFQKDVHDVLTNNTGTKTVEGLVLNLETTSRASFNTSAF*EMKKLR 361
Query: 553 LLQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYSSVKQ 606
LLQL+CVDLTGD+ LS +L+WV W+ T ++P++FY GNLV ELKY VKQ
Sbjct: 362 LLQLDCVDLTGDFGFLSKQLRWVNWRQSTFNHVPNNFYQGNLVXFELKYXMVKQ 523
>TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resistance gene
analog protein {Medicago ruthenica}, partial (98%)
Length = 1285
Score = 239 bits (611), Expect = 3e-63
Identities = 146/399 (36%), Positives = 230/399 (57%), Gaps = 8/399 (2%)
Frame = +3
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
GEDTR++F + L+AAL GI F DD L KG + P LLR IEGS++ +AVLS NYAS
Sbjct: 93 GEDTRNNFTNFLFAALERKGIYAFRDDTNLPKGESIGPELLRTIEGSQVFVAVLSRNYAS 272
Query: 78 SRWCLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQKG---PVLLLGKQLQVEDTQR-- 132
S WCL EL KI EC + G+ V+P+FY VDPS V+ Q G +Q +D +
Sbjct: 273 STWCLQELEKICECIKGSGKYVLPIFYGVDPSEVKKQSGIYWDDFAKHEQRFKQDPHKVS 452
Query: 133 KTRRGGHEVASFSGYNLSPDRSEAEAINEIVKNVFRNLDIRLMSITDFPVGLESHLQEV- 191
+ R ++V S +G++L D+ ++ + +IV+ + L + ++ VG+ S + +
Sbjct: 453 RWREALNQVGSIAGWDLR-DKQQSVEVEKIVQTILNILKCKSSFVSKDLVGINSRTEALK 629
Query: 192 IKLIESQSSKVCMVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCENDSRGHM 251
+L+ + V ++GIWGMGG GKTT A ++ QI +F + FI+++ ++ G +
Sbjct: 630 HQLLLNSVDGVRVIGIWGMGGKGKTTLAMNLYGQICHRFDASCFIDDVSKIFRLHD-GPI 806
Query: 252 HLQEQLLSDVLKTK-EKIHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALCGNREWF 310
Q+Q+L L + +I + T +I+ LS ++ L+ILD+V EQL + +REW
Sbjct: 807 DAQKQILHQTLGIEHHQICNHYSATDLIRHRLSREKTLLILDNVDQVEQLERIGVHREWL 986
Query: 311 ASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFG-EASPREDFNELSRR 369
+GS I++ +RD +L K D Y + +D ES +LF AF E +++ L+
Sbjct: 987 GAGSRIVIISRDEHILKEYKVDVDYKVPLLDWTESHKLFRQKAFKLEKIIMKNYQNLAYE 1166
Query: 370 VVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPN 408
++ Y GLPL + VLGS+L+ R EW+S L++L + PN
Sbjct: 1167ILNYANGLPLTITVLGSFLSGRNVTEWKSALARLRQRPN 1283
>BQ148669 similar to PIR|T07656|T076 probable resistance protein - soybean
(fragment), partial (63%)
Length = 608
Score = 138 bits (348), Expect(2) = 8e-62
Identities = 66/103 (64%), Positives = 82/103 (79%)
Frame = +2
Query: 356 EASPREDFNELSRRVVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKL 415
E+ + F ++S VV YCGGLPLALEVLGSYL +R+ +WE +L KL+RIPNDQVQ+KL
Sbjct: 8 ESESKRKFAQISINVV*YCGGLPLALEVLGSYLFDRQVTKWECLLEKLKRIPNDQVQKKL 187
Query: 416 RISYDGLKDELEKDIFLDICCFFVGKDRAYVTDILNGCGLYAD 458
+ISYDGL D+ E+DIFLDI FF+G DR V DILNGCGL+A+
Sbjct: 188 KISYDGLNDDTERDIFLDIAFFFIGMDRNDVMDILNGCGLFAE 316
Score = 118 bits (296), Expect(2) = 8e-62
Identities = 58/96 (60%), Positives = 74/96 (76%)
Frame = +3
Query: 460 GITLLIERSLLQVEKNNKLGMHDLVRDMGRAIVRESSTKDLGKRSRLWFHEDVHDVLTKN 519
GI++L+ERSL+ ++ NKLGMHDL+RDMGR I+R+ S K L KRSRLWFHEDVHDV
Sbjct: 321 GISVLVERSLVTIDDKNKLGMHDLLRDMGREIIRQKSPKKLEKRSRLWFHEDVHDVFVIT 500
Query: 520 TGTETVEGLILKLQSTSRVCFSTNAFKEMRKLRLLQ 555
+ GL+LKL + ++ CFSTNAF+ M+KLRLLQ
Sbjct: 501 KWCTSC*GLVLKLAANAK-CFSTNAFENMKKLRLLQ 605
>BQ148280 similar to GP|7107246|gb| unknown {Cicer arietinum}, partial (98%)
Length = 668
Score = 228 bits (582), Expect = 7e-60
Identities = 120/223 (53%), Positives = 160/223 (70%), Gaps = 2/223 (0%)
Frame = +1
Query: 214 GKTTTAKAIFNQIHRKFMCTSFIENIREVCENDSRGHMHLQEQLLSDVLKTKE-KIHSIG 272
GKTT AKA +N+I F SF+ N+RE E+D+ G + LQ++LLSD+ KT E KI ++
Sbjct: 1 GKTTIAKAAYNKIRHDFDAKSFLLNVREDWEHDN-GQVSLQQRLLSDIYKTTEIKIRTLE 177
Query: 273 RGTAMIKKILSGKRALVILDDVTTSEQLRALCGNREWFASGSVIIVTTRDVRLLNSLKAD 332
G ++K+ L K+ ++LDDV +QL ALCG+ EWF GS II+TTRD LL+ LK
Sbjct: 178 SGKMILKERLQKKKIFLVLDDVNKEDQLNALCGSHEWFGEGSRIIITTRDDDLLSRLKVH 357
Query: 333 YVYTMKEMDEDESLELFSWHAFGEASPREDFNELSRRVVAYCGGLPLALEVLGSY-LNER 391
YVY MKEMD++ESLELFSWHAF + +P + F LS VV Y GGLPLAL+V+GS+ L R
Sbjct: 358 YVYRMKEMDDNESLELFSWHAFKQPNPIKGFGNLSTDVVKYSGGLPLALQVIGSFLLTRR 537
Query: 392 EEIEWESVLSKLERIPNDQVQQKLRISYDGLKDELEKDIFLDI 434
+ EW S+L KL+ IPND+V +KL++S+DGL D+ K+IFLDI
Sbjct: 538 RKKEWTSLLEKLKLIPNDKVLEKLQLSFDGLSDDDMKEIFLDI 666
>TC89557 weakly similar to GP|10178211|dbj|BAB11635. TMV resistance protein N
{Arabidopsis thaliana}, partial (8%)
Length = 1567
Score = 102 bits (255), Expect(3) = 1e-58
Identities = 57/153 (37%), Positives = 91/153 (59%)
Frame = +3
Query: 286 RALVILDDVTTSEQLRALCGNREWFASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDES 345
+ L++LD+V EQL L ++ S II+ TRD +L + AD V+ K M+++++
Sbjct: 1110 KLLIVLDNVEQLEQLEKLDIEPKFLHPRSKIIIITRDKHILQAYGADEVFEAKLMNDEDA 1289
Query: 346 LELFSWHAFGEASPREDFNELSRRVVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLER 405
+L AF P F EL +V+ Y LPLA++VLGS+L R EW S L K E+
Sbjct: 1290 HKLLCRKAFKSDYPSSGFAELIPKVLVYAQRLPLAVKVLGSFLFSRNANEWSSTLDKFEK 1469
Query: 406 IPNDQVQQKLRISYDGLKDELEKDIFLDICCFF 438
P +++ + L++SY+GL+ + EK++FL + FF
Sbjct: 1470 NPPNKIMKALQVSYEGLEKD-EKEVFLHVAXFF 1565
Score = 89.7 bits (221), Expect(3) = 1e-58
Identities = 45/86 (52%), Positives = 56/86 (64%)
Frame = +3
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
G DTR+SFV HLYA L+ GI F DD++L+KG + P LL+AI+ SRIS+ V S +YAS
Sbjct: 288 GSDTRNSFVDHLYAHLNRKGIFTFKDDKQLQKGEAISPQLLQAIQQSRISIVVFSKDYAS 467
Query: 78 SRWCLDELVKILECRRTYGQLVIPLF 103
S WCLDE+ I E R P F
Sbjct: 468 STWCLDEMAAINESRINLKTSCFPCF 545
Score = 74.7 bits (182), Expect(3) = 1e-58
Identities = 47/156 (30%), Positives = 86/156 (55%), Gaps = 7/156 (4%)
Frame = +1
Query: 97 QLVIPLFYHVDPSHVRNQKGP---VLLLGKQLQVEDTQR--KTRRGGHEVASFSGYNLSP 151
Q+V P+FY VDPSHV+ Q G +L + ++ + + R + +G+++
Sbjct: 526 QVVFPVFYDVDPSHVKKQNGVYENAFVLHTEAFKHNSGKVARWRTTMTYLGGTAGWDVRV 705
Query: 152 DRSEAEAINEIVKNVFRNLDIRLMSITDFPVGLESHLQEVIKLIE--SQSSKVCMVGIWG 209
+ E E I +IV+ V + L TD +G++ H++ + L++ S+ ++GIWG
Sbjct: 706 E-PEFEMIEKIVEAVIKKLGRTFSGSTDDLIGIQPHIEALENLLKLSSKDDGCRVLGIWG 882
Query: 210 MGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCEN 245
M G+GKTT A ++++I +F FIEN+ ++ E+
Sbjct: 883 MDGIGKTTLATVLYDKISFQFDACCFIENVSKIYED 990
>TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (23%)
Length = 1651
Score = 221 bits (563), Expect = 1e-57
Identities = 128/343 (37%), Positives = 205/343 (59%), Gaps = 3/343 (0%)
Frame = +1
Query: 149 LSPDRSEAEAINEIVKNVFRNLDIRLMSITDFPVGLESHLQEVIKLIESQSS-KVCMVGI 207
L +R E + I +IV+++ N++ +++ +PVGL+S +++V L++ S +V MVG+
Sbjct: 484 LDTNRYEYKFIEKIVEDISNNINHVFLNVAKYPVGLQSRIEQVKLLLDMGSEDEVRMVGL 663
Query: 208 WGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCENDSRGHMHLQEQLLSDVLKTKEK 267
+G GG+GK+T AKA++N + +F F+ N+RE + + HLQ++LLS ++K K
Sbjct: 664 FGTGGMGKSTLAKAVYNFVADQFEGVCFLHNVRE--NSTHKNLKHLQKKLLSKIVKFDGK 837
Query: 268 IHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALCGNREWFASGSVIIVTTRDVRLLN 327
+ + G +IK+ LS K+ L+ILDDV EQL AL G +WF GS +I+TTRD LL
Sbjct: 838 LEDVSEGIPIIKERLSRKKILLILDDVDKLEQLEALAGGLDWFGHGSRVIITTRDKHLLA 1017
Query: 328 SLKADYVYTMKEMDEDESLELFSWHAFGEASPREDFNELSRRVVAYCGGLPLALEVLGSY 387
+ ++E++E E+LEL AF + E+ RVV Y GLPLA+ +G
Sbjct: 1018CHGITSTHAVEELNETEALELLRRMAFKNDKVPSTYEEILNRVVTYASGLPLAIVTIGDN 1197
Query: 388 LNEREEIEWESVLSKLERIPNDQVQQKLRISYDGLKDELEKDIFLDICCFFVGKDRAYVT 447
L R+ +W+ +L + E IPN +Q+ L++SYD L+ + EK +FLDI C F G V
Sbjct: 1198LFGRKVEDWKRILDEYENIPNKDIQRILQVSYDALEPK-EKSVFLDIACCFKGCKWTKVK 1374
Query: 448 DILNG-CGLYADIGITLLIERSLL-QVEKNNKLGMHDLVRDMG 488
IL+ G + + +L E+SL+ E + ++ +HDL+ DMG
Sbjct: 1375KILHAHYGHCIEHHVGVLAEKSLIGHWEYDTQMTLHDLIEDMG 1503
Score = 59.7 bits (143), Expect = 6e-09
Identities = 32/85 (37%), Positives = 50/85 (58%), Gaps = 8/85 (9%)
Frame = +2
Query: 75 YASSRWCLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQKGPV--LLLGKQLQVEDTQR 132
YASS +CLDELV I+ C +T LV+P+FY V+P+H+R+Q G L + + ++ ++
Sbjct: 2 YASSSFCLDELVHIIHCYKTKSCLVLPVFYDVEPTHIRHQSGSYGEYLTKHEERFQNNEK 181
Query: 133 KTRR------GGHEVASFSGYNLSP 151
R + A+ SGY+ SP
Sbjct: 182 NMERLRQWKIALTQAANLSGYHYSP 256
>BF643292 weakly similar to GP|22037340|gb| disease resistance-like protein
GS3-4 {Glycine max}, partial (45%)
Length = 576
Score = 214 bits (546), Expect(2) = 7e-56
Identities = 105/161 (65%), Positives = 125/161 (77%)
Frame = +1
Query: 347 ELFSWHAFGEASPREDFNELSRRVVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERI 406
E FSWHAF +A P +DF+E+S VV Y G LPLALEVLGSYL +RE EW VL KL+RI
Sbjct: 1 EFFSWHAFKQARPSKDFSEISTNVVQYSGRLPLALEVLGSYLFDREVTEWICVLEKLKRI 180
Query: 407 PNDQVQQKLRISYDGLKDELEKDIFLDICCFFVGKDRAYVTDILNGCGLYADIGITLLIE 466
PNDQV QKL+ISYDGL D+ EK IFLDI CFF+G DR V ILNG G +A+IGI++L+E
Sbjct: 181 PNDQVHQKLKISYDGLNDDTEKSIFLDIACFFIGMDRNDVIHILNGSGFFAEIGISVLVE 360
Query: 467 RSLLQVEKNNKLGMHDLVRDMGRAIVRESSTKDLGKRSRLW 507
RSL+ V+ NKLGMHDL+RDMGR I+RE S + +RSRLW
Sbjct: 361 RSLVTVDDKNKLGMHDLLRDMGREIIREKSPMEPEERSRLW 483
Score = 22.3 bits (46), Expect(2) = 7e-56
Identities = 9/17 (52%), Positives = 14/17 (81%)
Frame = +3
Query: 516 LTKNTGTETVEGLILKL 532
L+++TGT+ EGL LK+
Sbjct: 510 LSEHTGTKAGEGLTLKM 560
>BG448244 weakly similar to PIR|T08833|T088 disease resistance protein
homolog RLG6 - soybean (fragment), partial (34%)
Length = 599
Score = 213 bits (543), Expect = 2e-55
Identities = 104/169 (61%), Positives = 130/169 (76%)
Frame = +2
Query: 290 ILDDVTTSEQLRALCGNREWFASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELF 349
+LDDV+ EQ ALC GSVII+TTRD+R+LN L+ D++Y + ++ ESLELF
Sbjct: 95 VLDDVSELEQFNALCEGNS-VGPGSVIIITTRDLRVLNILEVDFIYEAEGLNASESLELF 271
Query: 350 SWHAFGEASPREDFNELSRRVVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPND 409
HAF + P EDF LSR VVAYCGG+PLALEVLGSYL +R + EW+SVLSKLE+IPND
Sbjct: 272 CGHAFRKVIPTEDFLILSRYVVAYCGGIPLALEVLGSYLLKRRKQEWQSVLSKLEKIPND 451
Query: 410 QVQQKLRISYDGLKDELEKDIFLDICCFFVGKDRAYVTDILNGCGLYAD 458
Q+ +KL+I ++GL D +E DIFLD+CCFF+GKDR YVT ILNG GL+AD
Sbjct: 452 QIHEKLKIXFNGLSDRMEXDIFLDVCCFFIGKDRXYVTKILNGXGLHAD 598
>TC86801 similar to PIR|A54810|A54810 TMV resistance protein N - tobacco
(Nicotiana glutinosa), partial (7%)
Length = 1268
Score = 208 bits (530), Expect = 8e-54
Identities = 132/341 (38%), Positives = 196/341 (56%), Gaps = 8/341 (2%)
Frame = +2
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
GEDTR F SHL+AALS ++ ++D +++KG E+ P L +AI+ S + L V S NYAS
Sbjct: 92 GEDTRAGFTSHLHAALSRTYLHTYID-YRIEKGDEVWPELEKAIKQSTLFLVVFSENYAS 268
Query: 78 SRWCLDELVKILECRRTYGQ---LVIPLFYHVDPSHVRNQK---GPVLLLGKQLQVEDTQ 131
S WCL+ELV+++ECR VIP+FYHVDPSHVR Q G L KQ +D
Sbjct: 269 STWCLNELVELMECRNKNEDDNIGVIPVFYHVDPSHVRKQTGSYGSALAKHKQENQDDKM 448
Query: 132 RKTRRGG-HEVASFSGYNLSPDRSEAEAINEIVKNVFRNLDIRLMSITDFPVGLESHLQE 190
+ + + A+ SG++ S R+E+ I +I + + L+ + + L+ +
Sbjct: 449 MQNWKNALFQAANLSGFHSSTYRTESNMIEDITRALLGKLNHQYRDELTCNLILDENYWA 628
Query: 191 VIKLIESQSSKVCMVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCENDSRGH 250
V LI+ S+ V ++G+WGMGG GKTT A A+F + K+ F+E + EV + G
Sbjct: 629 VRSLIKFDSTTVQIIGLWGMGGTGKTTLAAAMFQRFSFKYEGNCFLERVTEV--SKKHGI 802
Query: 251 MHLQEQLLSDVLKTKEKIHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALCGNR-EW 309
+ +LLS +L +I + AMIK+ L ++ ++LDDV SE L+ L G R W
Sbjct: 803 NYTCNKLLSKLLGEDLRIDTPKVIPAMIKRRLRHMKSFIVLDDVHNSELLQDLIGVRGGW 982
Query: 310 FASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFS 350
GS++IVTTRD +L S D +Y +K+M+ SL+LFS
Sbjct: 983 LGPGSIVIVTTRDKHVLISGGIDEIYEVKKMNSQNSLQLFS 1105
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,785,851
Number of Sequences: 36976
Number of extensions: 493922
Number of successful extensions: 3285
Number of sequences better than 10.0: 313
Number of HSP's better than 10.0 without gapping: 2968
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3082
length of query: 1069
length of database: 9,014,727
effective HSP length: 106
effective length of query: 963
effective length of database: 5,095,271
effective search space: 4906745973
effective search space used: 4906745973
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0342a.5


