

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0342a.1
(587 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
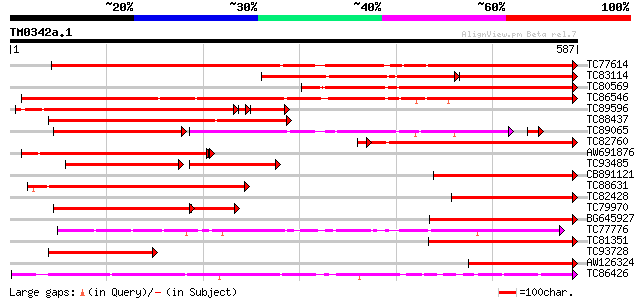
Score E
Sequences producing significant alignments: (bits) Value
TC77614 similar to PIR|JC5229|JC5229 laccase (EC 1.10.3.2) precu... 652 0.0
TC83114 similar to GP|1621467|gb|AAB17194.1| laccase {Liriodendr... 328 e-160
TC80569 similar to GP|9757923|dbj|BAB08370.1 laccase (diphenol o... 508 e-144
TC86546 weakly similar to PIR|T45944|T45944 laccase-like protein... 494 e-140
TC89596 similar to GP|9757923|dbj|BAB08370.1 laccase (diphenol o... 384 e-125
TC88437 similar to GP|1621463|gb|AAB17192.1| laccase {Liriodendr... 420 e-118
TC89065 weakly similar to GP|6478936|gb|AAF14041.1| putative lac... 256 e-115
TC82760 similar to GP|3805964|emb|CAA74105.1 laccase {Populus ba... 352 4e-98
AW691876 similar to GP|3805964|emb laccase {Populus balsamifera ... 305 1e-82
TC93485 similar to GP|21552583|gb|AAM54731.1 diphenol oxidase la... 182 3e-72
CB891121 similar to GP|1621467|gb| laccase {Liriodendron tulipif... 265 3e-71
TC88631 weakly similar to PIR|T00579|T00579 probable laccase [im... 262 2e-70
TC82428 similar to GP|1621461|gb|AAB17191.1| laccase {Liriodendr... 238 5e-63
TC79970 similar to GP|21552583|gb|AAM54731.1 diphenol oxidase la... 189 5e-60
BG645927 similar to GP|11071902|em laccase {Populus balsamifera ... 225 3e-59
TC77776 L-ascorbate oxidase 217 1e-56
TC81351 similar to PIR|T00579|T00579 probable laccase [imported]... 194 8e-50
TC93728 similar to GP|1621467|gb|AAB17194.1| laccase {Liriodendr... 193 2e-49
AW126324 similar to GP|20161818|db putative laccase {Oryza sativ... 189 3e-48
TC86426 similar to PIR|T07634|T07634 pollen-specific protein hom... 187 7e-48
>TC77614 similar to PIR|JC5229|JC5229 laccase (EC 1.10.3.2) precursor -
common tobacco, partial (96%)
Length = 2063
Score = 652 bits (1682), Expect = 0.0
Identities = 311/544 (57%), Positives = 393/544 (72%)
Frame = +2
Query: 44 QNVTRLCHSKSMVTVNGEFPGPHIVAREGDRLIIKVVNHVQNNISIHWHGIRQLQSGWAD 103
+N +RLC SK++VTVNG+FPGP + ARE D +I+KV N V NNI+IHWHGIRQL++GWAD
Sbjct: 152 KNTSRLCSSKAIVTVNGKFPGPTLYAREDDTVIVKVRNQVNNNITIHWHGIRQLRTGWAD 331
Query: 104 GPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRATLYGSLIILPKHNAQYPFVK 163
GPAY+TQCPIQ G SY YN+TI GQRGTL WHAH++WLR+T++G+++ILPK YPF K
Sbjct: 332 GPAYITQCPIQPGHSYTYNFTITGQRGTLLWHAHVNWLRSTVHGAIVILPKKGVPYPFPK 511
Query: 164 PYKEVPMIFGEWFNADPEAIISQALQTGGGPNVSDAYTINGLPGPLYNCSKKDTFKLKVK 223
P E+ ++ GEW+ +D EA+I++AL++G PNVSDA+TINGLPG + NCS +D +KL V+
Sbjct: 512 PDDELVLVLGEWWKSDTEAVINEALKSGLAPNVSDAHTINGLPGTVANCSTQDVYKLPVE 691
Query: 224 PGKIYLLRLINAALNDELFFSIANHTLKVVEADAVYVKPFVTKTILITPGQTTNVLLKTK 283
GK YLLR+INAALN+ELFF IA H L VVE DA Y KPF +TI+I PGQTTNVLL K
Sbjct: 692 SGKTYLLRIINAALNEELFFKIAGHKLTVVEVDATYTKPFQIETIVIAPGQTTNVLL--K 865
Query: 284 SHYPNATFFMTARPYVTGLGTFDNSTVAGILEYKIQHNITHHNSAVKLKKLPLFKPLLPA 343
++ + + + A P++ DN T L Y S L P F P
Sbjct: 866 ANQKSGKYLVAASPFMDAPVAVDNLTATATLHY----------SGTTLTNTPTFLTTPPP 1015
Query: 344 LNDTSFATKFSNKLRSLASARFPANVPQKVDKHFLFTVGLGTNPCKNKSNQTCQGPNNAT 403
N T A F N L+ L S ++P NVP K+D FTVGLG NPC +C+ N +
Sbjct: 1016TNATQIANNFLNSLKGLNSKKYPVNVPLKIDHSLFFTVGLGVNPC-----PSCKAGNGSR 1180
Query: 404 MFSGSVNNVSFILPTTALLQAHFFGKSNGVYTPDFPTSPLHPFNYTGTTPNNTNVSNGTK 463
+ + ++NNV+F++PTTALLQAH+F N V+T DFP +P H +N+TG P N N ++GTK
Sbjct: 1181VVA-AINNVTFVMPTTALLQAHYFNIKN-VFTADFPPNPPHIYNFTGAGPKNLNTTSGTK 1354
Query: 464 VVVLPFNTSVELVMQDTSILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSNFNLVDPVE 523
+ L FN +V+LVMQDT I+ ESHP+HLHGFNFFVVG+G GN+D D FNLVDPVE
Sbjct: 1355LYKLSFNDTVQLVMQDTGIIAPESHPVHLHGFNFFVVGRGVGNYDSKNDSKKFNLVDPVE 1534
Query: 524 RNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTSWGLKMAWVVLDGKLPNQKLLPPPVD 583
RNTVGVP+GGWVAIRF ADNPGVWFMHCHLEVHT+WGLKMA++V +GK P Q ++ PP D
Sbjct: 1535RNTVGVPAGGWVAIRFRADNPGVWFMHCHLEVHTTWGLKMAFLVDNGKGPKQSVIAPPKD 1714
Query: 584 LPKC 587
LPKC
Sbjct: 1715LPKC 1726
>TC83114 similar to GP|1621467|gb|AAB17194.1| laccase {Liriodendron
tulipifera}, partial (54%)
Length = 1216
Score = 328 bits (841), Expect(2) = e-160
Identities = 161/205 (78%), Positives = 177/205 (85%)
Frame = +1
Query: 261 KPFVTKTILITPGQTTNVLLKTKSHYPNATFFMTARPYVTGLGTFDNSTVAGILEYKIQH 320
KPFVT TILI PGQTTNVLLKTK HYPNATF M ARPY TG GTFDNSTVAGI+EY+I
Sbjct: 4 KPFVTNTILIAPGQTTNVLLKTKPHYPNATFLMLARPYATGQGTFDNSTVAGIIEYEIPF 183
Query: 321 NITHHNSAVKLKKLPLFKPLLPALNDTSFATKFSNKLRSLASARFPANVPQKVDKHFLFT 380
N H NS+ LKKLPL KP+LP LNDTSFAT F+NKL SLA+A+FPANVPQKVDKHF FT
Sbjct: 184 NTHHSNSS--LKKLPLLKPILPQLNDTSFATNFTNKLHSLANAQFPANVPQKVDKHFFFT 357
Query: 381 VGLGTNPCKNKSNQTCQGPNNATMFSGSVNNVSFILPTTALLQAHFFGKSNGVYTPDFPT 440
VGLGTNPC+NK NQTCQGP N TMF+ SVNNVSFI+PTTALLQ HFFG++NG+YT DFP+
Sbjct: 358 VGLGTNPCQNK-NQTCQGP-NGTMFAASVNNVSFIMPTTALLQTHFFGQNNGIYTTDFPS 531
Query: 441 SPLHPFNYTGTTPNNTNVSNGTKVV 465
P++PFNYTGT PNNT VSNGTKVV
Sbjct: 532 KPMNPFNYTGTPPNNTMVSNGTKVV 606
Score = 254 bits (650), Expect(2) = e-160
Identities = 114/122 (93%), Positives = 118/122 (96%)
Frame = +2
Query: 466 VLPFNTSVELVMQDTSILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSNFNLVDPVERN 525
VLPFNTSVELV+QDTSILG ESHPLHLHGFNFFVVGQGFGNFD N DP NFNLVDPVERN
Sbjct: 608 VLPFNTSVELVLQDTSILGVESHPLHLHGFNFFVVGQGFGNFDSNSDPQNFNLVDPVERN 787
Query: 526 TVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTSWGLKMAWVVLDGKLPNQKLLPPPVDLP 585
TVGVPSGGWVAIRFLADNPGVWFMHCHLE+HTSWGLKMAW+VLDGKLPNQK+LPPPVDLP
Sbjct: 788 TVGVPSGGWVAIRFLADNPGVWFMHCHLEIHTSWGLKMAWIVLDGKLPNQKVLPPPVDLP 967
Query: 586 KC 587
KC
Sbjct: 968 KC 973
>TC80569 similar to GP|9757923|dbj|BAB08370.1 laccase (diphenol oxidase)
{Arabidopsis thaliana}, partial (46%)
Length = 1117
Score = 508 bits (1308), Expect = e-144
Identities = 246/285 (86%), Positives = 259/285 (90%)
Frame = +1
Query: 303 GTFDNSTVAGILEYKIQHNITHHNSAVKLKKLPLFKPLLPALNDTSFATKFSNKLRSLAS 362
GTFDN+TVAGILEY+I N THH SA LKK+PLFKP LPALNDTSFATKFS KLRSLAS
Sbjct: 4 GTFDNTTVAGILEYEIPSN-THH-SASSLKKIPLFKPTLPALNDTSFATKFSKKLRSLAS 177
Query: 363 ARFPANVPQKVDKHFLFTVGLGTNPCKNKSNQTCQGPNNATMFSGSVNNVSFILPTTALL 422
+FPANVPQKVDKHF FTVGLGTNPC+ SNQTCQGPN TMF+ SVNNVSF +PTTALL
Sbjct: 178 PQFPANVPQKVDKHFFFTVGLGTNPCQ--SNQTCQGPNG-TMFAASVNNVSFTMPTTALL 348
Query: 423 QAHFFGKSNGVYTPDFPTSPLHPFNYTGTTPNNTNVSNGTKVVVLPFNTSVELVMQDTSI 482
Q+HF G+S GVY P FP+SPLHPFNYTGT PNNT VSNGTKV VLPFNTSVELVMQDTSI
Sbjct: 349 QSHFTGQSRGVYAPYFPSSPLHPFNYTGTPPNNTMVSNGTKVTVLPFNTSVELVMQDTSI 528
Query: 483 LGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSNFNLVDPVERNTVGVPSGGWVAIRFLAD 542
LGAESHPLHLHGFNFFVVGQGFGN+D NKDP NFNLVDPVERNT+GVPSGGWVAIRFLAD
Sbjct: 529 LGAESHPLHLHGFNFFVVGQGFGNYDSNKDPKNFNLVDPVERNTIGVPSGGWVAIRFLAD 708
Query: 543 NPGVWFMHCHLEVHTSWGLKMAWVVLDGKLPNQKLLPPPVDLPKC 587
NPGVWFMHCHLEVHTSWGLKMAW+VLDGKLPNQKLLPPP DLPKC
Sbjct: 709 NPGVWFMHCHLEVHTSWGLKMAWIVLDGKLPNQKLLPPPADLPKC 843
>TC86546 weakly similar to PIR|T45944|T45944 laccase-like protein -
Arabidopsis thaliana, partial (64%)
Length = 2066
Score = 494 bits (1273), Expect = e-140
Identities = 252/584 (43%), Positives = 358/584 (61%), Gaps = 9/584 (1%)
Frame = +3
Query: 13 LYLFISLCYILFYSFVCSSCNKIRLLTQIRYQNVTRLCHSKSMVTVNGEFPGPHIVAREG 72
+ L+ + + L S +S + + + + RLC+ + +VTVNG +PGP + R+G
Sbjct: 36 ILLWQTWAFALILSSFMASAATVEHTFLVENKTIKRLCNEQVIVTVNGLYPGPKLEVRDG 215
Query: 73 DRLIIKVVNHVQNNISIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTL 132
D +I+ V+N+ NI+IHWHG+ QL S W+DGP Y+TQC I+ + Y + + Q GTL
Sbjct: 216 DSVIVHVINNSPYNITIHWHGVFQLYSAWSDGPEYITQCSIRPENKFTYKFNVTQQEGTL 395
Query: 133 FWHAHISWLRATLYGSLIILPKHNAQYPFVKPYKEVPMIFGEWFNADPEAIISQALQTGG 192
+WHAH S +RAT++G++II P+ + ++PF KPYKEVP+I G+W++ + E II + L+TG
Sbjct: 396 WWHAHASVVRATVHGAIIIQPR-SGRFPFPKPYKEVPIILGDWYDGNVEEIIQKELETGD 572
Query: 193 GPNVSDAYTINGLPGPLYNCSKKDTFKLKVKPGKIYLLRLINAALNDELFFSIANHTLKV 252
SDA+TING PG L+NCSK +KLKVK GK YL R++NAAL + LFF IA+H V
Sbjct: 573 -KIASDAFTINGFPGDLFNCSKNQMYKLKVKQGKTYLFRMVNAALANNLFFKIADHKFTV 749
Query: 253 VEADAVYVKPFVTKTILITPGQTTNVLLKTKSHYPNATFFMTARPYVTG--LGTFDNSTV 310
V DA Y +P+ T I+I GQ+ ++L P +++M A PYV G +G FDNST
Sbjct: 750 VAMDAAYTEPYTTDIIVIAAGQSADILFTADQ--PKGSYYMAASPYVVGEPVGLFDNSTT 923
Query: 311 AGILEYKIQHNITHHNSAVKLKKLPLFKPLLPALNDTSFATKFSNKLRSLASARFPANVP 370
++ Y+ + K K + P LP N+T A KF + + L VP
Sbjct: 924 RAVVFYEGYKKL-------KTKHIVPLMPALPLHNNTPIAHKFFSNITGLVGGPNWVPVP 1082
Query: 371 QKVDKHFLFTVGLGTNPCKNKSNQTCQGPNNATMFSGSVNNVSFILPTT---ALLQAHFF 427
+VD+H T+ +G PC N C GP F+ S+NN SF+LP ++++A+F+
Sbjct: 1083LEVDEHMYITINMGLVPCP--VNAKCTGPLGQK-FASSMNNESFLLPVGKGYSIMEAYFY 1253
Query: 428 GKSNGVYTPDFPTSPLHPFNYTGTT----PNNTNVSNGTKVVVLPFNTSVELVMQDTSIL 483
S G+YT DFP +P F++ PN T TKV +N++VE+V Q+T+IL
Sbjct: 1254NVS-GIYTTDFPDNPPKFFDFVNPKIFLDPNVTFTPKSTKVKQFKYNSTVEIVFQNTAIL 1430
Query: 484 GAESHPLHLHGFNFFVVGQGFGNFDPNKDPSNFNLVDPVERNTVGVPSGGWVAIRFLADN 543
A+SHP+HLHG NF V+ Q FG F+P D +NLV+P RNTV VP GGW AIRF +N
Sbjct: 1431NAQSHPMHLHGMNFHVLAQDFGIFNPTTDKLKYNLVNPSIRNTVAVPVGGWAAIRFRTNN 1610
Query: 544 PGVWFMHCHLEVHTSWGLKMAWVVLDGKLPNQKLLPPPVDLPKC 587
PGVWF+HCH++ H WGL A++V +G P+ L PPP DLPKC
Sbjct: 1611PGVWFLHCHVDDHNLWGLVTAFIVENGPTPSTSLGPPPADLPKC 1742
>TC89596 similar to GP|9757923|dbj|BAB08370.1 laccase (diphenol oxidase)
{Arabidopsis thaliana}, partial (44%)
Length = 921
Score = 384 bits (986), Expect(3) = e-125
Identities = 187/231 (80%), Positives = 200/231 (85%)
Frame = +2
Query: 7 LKIYTLLYLFISLCYILFYSFVCSSCNKIRLLTQIRYQNVTRLCHSKSMVTVNGEFPGPH 66
LKI TL F+SL + F + + I YQNVTRLCH+K MVTVNG+FPGP
Sbjct: 83 LKIATL---FLSLINFCAFEFALAGTTR-HYQFDIGYQNVTRLCHNKRMVTVNGQFPGPR 250
Query: 67 IVAREGDRLIIKVVNHVQNNISIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIK 126
I+AREGDRL+IKVVN+VQNNISIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIK
Sbjct: 251 IMAREGDRLVIKVVNNVQNNISIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIK 430
Query: 127 GQRGTLFWHAHISWLRATLYGSLIILPKHNAQYPFVKPYKEVPMIFGEWFNADPEAIISQ 186
GQRGTLFWHAHISWLR+TLYG LIILPK N YPF KP+KEVPMIFGEWFNAD EAII+Q
Sbjct: 431 GQRGTLFWHAHISWLRSTLYGPLIILPKKNVPYPFAKPHKEVPMIFGEWFNADTEAIIAQ 610
Query: 187 ALQTGGGPNVSDAYTINGLPGPLYNCSKKDTFKLKVKPGKIYLLRLINAAL 237
ALQTGGGPNVS+AYTINGLPGPLYNCSKKDTFKLKVKPGK YLLRLINAAL
Sbjct: 611 ALQTGGGPNVSEAYTINGLPGPLYNCSKKDTFKLKVKPGKTYLLRLINAAL 763
Score = 75.9 bits (185), Expect(3) = e-125
Identities = 36/40 (90%), Positives = 37/40 (92%)
Frame = +1
Query: 250 LKVVEADAVYVKPFVTKTILITPGQTTNVLLKTKSHYPNA 289
LKVVEADA+YVKPF T TILI PGQTTNVLLKTKSHYPNA
Sbjct: 802 LKVVEADAIYVKPFETNTILIAPGQTTNVLLKTKSHYPNA 921
Score = 28.9 bits (63), Expect(3) = e-125
Identities = 11/12 (91%), Positives = 12/12 (99%)
Frame = +3
Query: 238 NDELFFSIANHT 249
NDELFFS+ANHT
Sbjct: 765 NDELFFSVANHT 800
>TC88437 similar to GP|1621463|gb|AAB17192.1| laccase {Liriodendron
tulipifera}, partial (41%)
Length = 984
Score = 420 bits (1080), Expect = e-118
Identities = 195/252 (77%), Positives = 222/252 (87%), Gaps = 1/252 (0%)
Frame = +1
Query: 41 IRYQNVTRLCHSKSMVTVNGEFPGPHIVAREGDRLIIKVVNHVQNNISIHWHGIRQLQSG 100
IR NVTRLCH+KSMVTVNG+FPGP IV REGDRL++KVVNHV NNIS+HWHG+RQL+SG
Sbjct: 211 IRLANVTRLCHTKSMVTVNGKFPGPRIVVREGDRLLVKVVNHVPNNISLHWHGVRQLRSG 390
Query: 101 WADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRATLYGSLIILPKHNAQYP 160
W+DGP+Y+TQCPIQTGQSYVYN+TI GQRGTLFWHAH SWLRAT+YG LI+LP+HN YP
Sbjct: 391 WSDGPSYITQCPIQTGQSYVYNFTIVGQRGTLFWHAHFSWLRATVYGPLILLPRHNESYP 570
Query: 161 FVKPYKEVPMIFGEWFNADP-EAIISQALQTGGGPNVSDAYTINGLPGPLYNCSKKDTFK 219
F KPYKEVP++FGEW+ + + QTGGGPNVSDAYTINGLPGPLYNCS KDT+K
Sbjct: 571 FQKPYKEVPILFGEWWECRS*SCNLHKLFQTGGGPNVSDAYTINGLPGPLYNCS-KDTYK 747
Query: 220 LKVKPGKIYLLRLINAALNDELFFSIANHTLKVVEADAVYVKPFVTKTILITPGQTTNVL 279
LKVKPGK YLLRLINAALNDELFFSIANHTL +VEADA Y+KPF + TI++ PGQTTNVL
Sbjct: 748 LKVKPGKTYLLRLINAALNDELFFSIANHTLIIVEADASYIKPFESNTIILGPGQTTNVL 927
Query: 280 LKTKSHYPNATF 291
LKTK +YPN+TF
Sbjct: 928 LKTKPNYPNSTF 963
>TC89065 weakly similar to GP|6478936|gb|AAF14041.1| putative laccase
{Arabidopsis thaliana}, partial (63%)
Length = 1647
Score = 256 bits (655), Expect(3) = e-115
Identities = 145/345 (42%), Positives = 198/345 (57%), Gaps = 10/345 (2%)
Frame = +1
Query: 187 ALQTGGGPNVSDAYTINGLPGPLYNCSKKDTFKLKVKPGKIYLLRLINAALNDELFFSIA 246
A TG P S+A TINGLP LYNCS+ T+K+KVK GK YLLR+INAALN++ FF IA
Sbjct: 550 ATDTGNPPEESNASTINGLPSDLYNCSQDQTYKVKVKQGKTYLLRIINAALNEQHFFKIA 729
Query: 247 NHTLKVVEADAVYVKPFVTKTILITPGQTTNVLLKTKSHYPNATFFMTARPYVTGLGTFD 306
NH+ VV DA+Y + + T +++ PGQT +VLLKT +++M PY +
Sbjct: 730 NHSFTVVAMDAIYTEHYNTDVVVLAPGQTVDVLLKTNQVVD--SYYMVFTPYRSSNVGTS 903
Query: 307 NSTVAGILEYKIQHNITHHNSAVKLKKLPLFKPLLPALNDTSFATKFSNKLRSLASARFP 366
N T G++ Y K P+ P++P +DT A KF + L +
Sbjct: 904 NITTRGVVVY----------DGASQTKAPIM-PIMPDAHDTPTAHKFYTNVTGLTTGPHW 1050
Query: 367 ANVPQKVDKHFLFTVGLGTNPCKNKSNQTCQGPNNATMFSGSVNNVSFILPT---TALLQ 423
VP+KVD+H T G+G C N C N + S ++NN SF+LP +LL+
Sbjct: 1051VPVPRKVDEHMFITFGIGLEQCINPGPGRCVVLN--SRLSANMNNESFVLPKGRGFSLLE 1224
Query: 424 AHFFGKSNGVYTPDFPTSPLHPFNYTGTTPNNTNVS-------NGTKVVVLPFNTSVELV 476
A F+ +GVYT DFP P FNYT + N N S TKV L FN++VE+V
Sbjct: 1225A-FYKNISGVYTKDFPNQPPFEFNYTDPSLANVNPSEPLAFAPKSTKVKTLKFNSTVEIV 1401
Query: 477 MQDTSILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSNFNLVDP 521
+Q+T+ILG E+HP+H+HGFNF V+ QGFGN++ +D FN V+P
Sbjct: 1402LQNTAILGTENHPIHIHGFNFHVLAQGFGNYNATRDEPKFNFVNP 1536
Score = 166 bits (421), Expect(3) = e-115
Identities = 74/138 (53%), Positives = 98/138 (70%)
Frame = +3
Query: 46 VTRLCHSKSMVTVNGEFPGPHIVAREGDRLIIKVVNHVQNNISIHWHGIRQLQSGWADGP 105
+ RLC + + VNG PGP I AREGD +II V N N+++HWHGI Q + W+DGP
Sbjct: 126 IQRLCLPQVITAVNGTLPGPTINAREGDTVIIHVFNKSPYNLTLHWHGIIQFLTPWSDGP 305
Query: 106 AYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRATLYGSLIILPKHNAQYPFVKPY 165
YVTQCPI +G SY Y + + GQ GTL+WHAH S+LRAT++G+LII P+ YPF K Y
Sbjct: 306 EYVTQCPIPSGGSYTYQFNLTGQEGTLWWHAHSSFLRATVHGALIIKPRLGRSYPFPKVY 485
Query: 166 KEVPMIFGEWFNADPEAI 183
+EVP++ GEW+NA+ E +
Sbjct: 486 QEVPILLGEWWNANVEEV 539
Score = 33.9 bits (76), Expect(3) = e-115
Identities = 10/16 (62%), Positives = 14/16 (87%)
Frame = +3
Query: 537 IRFLADNPGVWFMHCH 552
+RF A+NPG+W +HCH
Sbjct: 1584 VRFQANNPGIWLVHCH 1631
>TC82760 similar to GP|3805964|emb|CAA74105.1 laccase {Populus balsamifera
subsp. trichocarpa}, partial (34%)
Length = 970
Score = 352 bits (903), Expect(2) = 4e-98
Identities = 163/219 (74%), Positives = 188/219 (85%), Gaps = 2/219 (0%)
Frame = +3
Query: 371 QKVDKHFLFTVGLGTNPCKNKSNQTCQGPNNATMFSGSVNNVSFILPTTALLQAHFFGKS 430
+K+ F FTVGLGT PC N TCQGPNN T F+ SVNN SF+LP+ +++QA++FGKS
Sbjct: 33 KKLTIKFFFTVGLGTFPCPK--NSTCQGPNNNTKFAASVNNFSFVLPSVSIMQAYYFGKS 206
Query: 431 N--GVYTPDFPTSPLHPFNYTGTTPNNTNVSNGTKVVVLPFNTSVELVMQDTSILGAESH 488
N GVY DFP +PL+PFNYTGT+PNNT V+N TK+VVL FNTSVELV+QDTSILGAESH
Sbjct: 207 NSNGVYKTDFPETPLNPFNYTGTSPNNTMVNNDTKLVVLNFNTSVELVLQDTSILGAESH 386
Query: 489 PLHLHGFNFFVVGQGFGNFDPNKDPSNFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWF 548
PLHLHG++FFVVGQGFGN+D NKDP+ FNLVDPVERNTVGVP+GGWVAIRF ADNPGVWF
Sbjct: 387 PLHLHGYDFFVVGQGFGNYDANKDPAKFNLVDPVERNTVGVPAGGWVAIRFFADNPGVWF 566
Query: 549 MHCHLEVHTSWGLKMAWVVLDGKLPNQKLLPPPVDLPKC 587
MHCHL++HTSWGL+MAW+VLDG NQKL PPP DLPKC
Sbjct: 567 MHCHLDIHTSWGLRMAWLVLDGPDSNQKLQPPPSDLPKC 683
Score = 25.0 bits (53), Expect(2) = 4e-98
Identities = 9/14 (64%), Positives = 13/14 (92%)
Frame = +2
Query: 361 ASARFPANVPQKVD 374
A+++FP NVP+KVD
Sbjct: 2 ANSKFPINVPKKVD 43
>AW691876 similar to GP|3805964|emb laccase {Populus balsamifera subsp.
trichocarpa}, partial (30%)
Length = 659
Score = 305 bits (780), Expect(2) = 1e-82
Identities = 135/197 (68%), Positives = 164/197 (82%)
Frame = +1
Query: 13 LYLFISLCYILFYSFVCSSCNKIRLLTQIRYQNVTRLCHSKSMVTVNGEFPGPHIVAREG 72
++LF ILF V C I Y+NVTRLCH++++++VNG+FPGP +VAREG
Sbjct: 46 VFLFGLCVIILFPELVV--CRTRHYTFNIEYKNVTRLCHTRTILSVNGKFPGPRLVAREG 219
Query: 73 DRLIIKVVNHVQNNISIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTL 132
DR+++KVVNH+ NN++IHWHGIRQ +GW+DGPAYVTQCPIQT Q+Y YN+TI GQRGTL
Sbjct: 220 DRVLVKVVNHISNNVTIHWHGIRQKTTGWSDGPAYVTQCPIQTNQTYTYNFTITGQRGTL 399
Query: 133 FWHAHISWLRATLYGSLIILPKHNAQYPFVKPYKEVPMIFGEWFNADPEAIISQALQTGG 192
FWHAHISWLRATLYG +IILPKHN YPF KP+KE+P++FGEWFN DPEA+I+QALQTGG
Sbjct: 400 FWHAHISWLRATLYGPIIILPKHNESYPFQKPHKEIPILFGEWFNVDPEAVINQALQTGG 579
Query: 193 GPNVSDAYTINGLPGPL 209
GPNVSDAYTINGLPG +
Sbjct: 580 GPNVSDAYTINGLPGTI 630
Score = 20.8 bits (42), Expect(2) = 1e-82
Identities = 6/7 (85%), Positives = 6/7 (85%)
Frame = +2
Query: 206 PGPLYNC 212
PGP YNC
Sbjct: 620 PGPFYNC 640
>TC93485 similar to GP|21552583|gb|AAM54731.1 diphenol oxidase laccase
{Glycine max}, partial (40%)
Length = 740
Score = 182 bits (461), Expect(2) = 3e-72
Identities = 73/123 (59%), Positives = 98/123 (79%)
Frame = +3
Query: 58 VNGEFPGPHIVAREGDRLIIKVVNHVQNNISIHWHGIRQLQSGWADGPAYVTQCPIQTGQ 117
VNG+FPGP + GD L++KV+N + N++IHWHG+RQ+++GWADGP +VTQCPI+ G+
Sbjct: 48 VNGQFPGPTLEINNGDTLVVKVINKARYNVTIHWHGVRQIRTGWADGPEFVTQCPIRPGE 227
Query: 118 SYVYNYTIKGQRGTLFWHAHISWLRATLYGSLIILPKHNAQYPFVKPYKEVPMIFGEWFN 177
SY Y +TI GQ GTL+WHAH SWLRAT+YG+LII PK YPF KP +E P++ GEW++
Sbjct: 228 SYTYRFTINGQEGTLWWHAHSSWLRATVYGALIIHPKEGDAYPFTKPKRETPILLGEWWD 407
Query: 178 ADP 180
A+P
Sbjct: 408 ANP 416
Score = 108 bits (271), Expect(2) = 3e-72
Identities = 53/94 (56%), Positives = 65/94 (68%)
Frame = +2
Query: 187 ALQTGGGPNVSDAYTINGLPGPLYNCSKKDTFKLKVKPGKIYLLRLINAALNDELFFSIA 246
A QTG PN+SDAYTING PG LY CS K T + + G+ L+R+INAALN L F+IA
Sbjct: 437 ATQTGAAPNISDAYTINGQPGDLYKCSSKGTTIVPIXSGETNLIRVINAALNQPLXFTIA 616
Query: 247 NHTLKVVEADAVYVKPFVTKTILITPGQTTNVLL 280
NH L ADA YVKPF T +++ P QTT+VL+
Sbjct: 617 NHKLTXXGADASYVKPFTTNVLMLGPDQTTDVLI 718
>CB891121 similar to GP|1621467|gb| laccase {Liriodendron tulipifera},
partial (25%)
Length = 706
Score = 265 bits (678), Expect = 3e-71
Identities = 120/149 (80%), Positives = 130/149 (86%)
Frame = -3
Query: 439 PTSPLHPFNYTGTTPNNTNVSNGTKVVVLPFNTSVELVMQDTSILGAESHPLHLHGFNFF 498
P++P FNYTGT P N V +GTKV VLP+NT VELV+QDTSILGAESHPLHLHGFNFF
Sbjct: 704 PSNPPVKFNYTGTPPKNIMVKSGTKVAVLPYNTKVELVLQDTSILGAESHPLHLHGFNFF 525
Query: 499 VVGQGFGNFDPNKDPSNFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTS 558
+VGQG GNFDP KDP+ FNLVDPVERNT GVP+GGWVA+RFLADNPGVWFMHCHLEVHTS
Sbjct: 524 IVGQGNGNFDPKKDPAKFNLVDPVERNTAGVPAGGWVALRFLADNPGVWFMHCHLEVHTS 345
Query: 559 WGLKMAWVVLDGKLPNQKLLPPPVDLPKC 587
WGLKMAW+V DGK NQKL PPP DLPKC
Sbjct: 344 WGLKMAWIVQDGKRRNQKLPPPPSDLPKC 258
>TC88631 weakly similar to PIR|T00579|T00579 probable laccase [imported] -
Arabidopsis thaliana, partial (38%)
Length = 779
Score = 262 bits (670), Expect = 2e-70
Identities = 127/235 (54%), Positives = 162/235 (68%), Gaps = 5/235 (2%)
Frame = +3
Query: 19 LCYIL-----FYSFVCSSCNKIRLLTQIRYQNVTRLCHSKSMVTVNGEFPGPHIVAREGD 73
LCY+L SF ++ N I+ V RLC ++ ++TVNG+FPGP I AR+GD
Sbjct: 39 LCYLLGLLTIIVSFASAAENHYHQFV-IQTATVKRLCKTRRILTVNGQFPGPTIEARDGD 215
Query: 74 RLIIKVVNHVQNNISIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLF 133
++IKV N NISIHWHG R L++ WADGP+YVTQCPIQ G SY Y +TI+ Q GTL+
Sbjct: 216 SMVIKVTNAGPYNISIHWHGFRMLRNPWADGPSYVTQCPIQPGGSYTYRFTIQNQEGTLW 395
Query: 134 WHAHISWLRATLYGSLIILPKHNAQYPFVKPYKEVPMIFGEWFNADPEAIISQALQTGGG 193
WHAH +LRAT+YG+ II PK + YPF P +E P++ GEWF+ DP A++ Q TG
Sbjct: 396 WHAHTGFLRATVYGAFIIYPKMGSPYPFSMPTREFPILLGEWFDRDPMALLRQTQFTGAP 575
Query: 194 PNVSDAYTINGLPGPLYNCSKKDTFKLKVKPGKIYLLRLINAALNDELFFSIANH 248
PNVS AYT+NG PG LY CS + T + +V G+ LLR+IN+ALN ELFFSIANH
Sbjct: 576 PNVSVAYTMNGQPGDLYRCSSQGTVRFQVYAGETILLRIINSALNQELFFSIANH 740
>TC82428 similar to GP|1621461|gb|AAB17191.1| laccase {Liriodendron
tulipifera}, partial (22%)
Length = 707
Score = 238 bits (607), Expect = 5e-63
Identities = 105/130 (80%), Positives = 118/130 (90%)
Frame = +1
Query: 458 VSNGTKVVVLPFNTSVELVMQDTSILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSNFN 517
VSNGTK VV+P+NT V++++QDTSILGAESHPLHLHGFNFFVVGQGFGNF+ + DP+ FN
Sbjct: 4 VSNGTKTVVIPYNTRVQVILQDTSILGAESHPLHLHGFNFFVVGQGFGNFNASSDPAKFN 183
Query: 518 LVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTSWGLKMAWVVLDGKLPNQKL 577
LVDPVERNTV VPSGGWVAIRFLADNPGVW MHCH +VH SWGL+MAW+V DGKLP+QKL
Sbjct: 184 LVDPVERNTVAVPSGGWVAIRFLADNPGVWLMHCHFDVHLSWGLRMAWIVEDGKLPDQKL 363
Query: 578 LPPPVDLPKC 587
PPP DLPKC
Sbjct: 364 PPPPKDLPKC 393
>TC79970 similar to GP|21552583|gb|AAM54731.1 diphenol oxidase laccase
{Glycine max}, partial (36%)
Length = 800
Score = 189 bits (479), Expect(2) = 5e-60
Identities = 77/146 (52%), Positives = 108/146 (73%)
Frame = +1
Query: 46 VTRLCHSKSMVTVNGEFPGPHIVAREGDRLIIKVVNHVQNNISIHWHGIRQLQSGWADGP 105
V RLC + + +TVNG++PGP + GD L++KV N + N++IHWHG+RQ+++GWADGP
Sbjct: 214 VKRLCKTHNTITVNGQYPGPTLEINNGDTLVVKVTNKARYNVTIHWHGVRQMRTGWADGP 393
Query: 106 AYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRATLYGSLIILPKHNAQYPFVKPY 165
+VTQCPI+ G SY Y +T+ GQ GTL+WHAH SWLRAT+YG+LII P+ YPF KP
Sbjct: 394 EFVTQCPIRPGGSYTYRFTVNGQEGTLWWHAHSSWLRATVYGALIIRPREGEPYPFPKPN 573
Query: 166 KEVPMIFGEWFNADPEAIISQALQTG 191
E ++ GEW++ +P ++ QA +TG
Sbjct: 574 HETSILLGEWWDGNPINVVRQAQRTG 651
Score = 61.2 bits (147), Expect(2) = 5e-60
Identities = 28/50 (56%), Positives = 35/50 (70%)
Frame = +2
Query: 189 QTGGGPNVSDAYTINGLPGPLYNCSKKDTFKLKVKPGKIYLLRLINAALN 238
+ G PN+SDAYTING PG LY CS K T + + G+ L+R+INAALN
Sbjct: 644 ELGAAPNISDAYTINGQPGDLYKCSTKGTTIIPIHSGETNLVRVINAALN 793
>BG645927 similar to GP|11071902|em laccase {Populus balsamifera subsp.
trichocarpa}, partial (38%)
Length = 574
Score = 225 bits (574), Expect = 3e-59
Identities = 101/153 (66%), Positives = 123/153 (80%)
Frame = -1
Query: 435 TPDFPTSPLHPFNYTGTTPNNTNVSNGTKVVVLPFNTSVELVMQDTSILGAESHPLHLHG 494
T DFP +P +N+TG+ N + GT++ L +N++V+LV+QDT +L E+HP+HLHG
Sbjct: 574 TDDFPGNPPVVYNFTGSQVTNLATTKGTRLYRLAYNSTVQLVLQDTGMLTPENHPIHLHG 395
Query: 495 FNFFVVGQGFGNFDPNKDPSNFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCHLE 554
FNFFVVG+G GNFD KD FNLVDPVERNTVGVP+GGW AIRF ADNPGVWFMHCHLE
Sbjct: 394 FNFFVVGRGQGNFDSKKDVKKFNLVDPVERNTVGVPAGGWTAIRFKADNPGVWFMHCHLE 215
Query: 555 VHTSWGLKMAWVVLDGKLPNQKLLPPPVDLPKC 587
+HT+WGLKMA+VV +GK PN+ LLPPP DLPKC
Sbjct: 214 IHTTWGLKMAFVVDNGKGPNESLLPPPSDLPKC 116
>TC77776 L-ascorbate oxidase
Length = 2066
Score = 217 bits (552), Expect = 1e-56
Identities = 179/557 (32%), Positives = 253/557 (45%), Gaps = 32/557 (5%)
Frame = +3
Query: 50 CHSKSMVTVNGEFPGPHIVAREGDRLIIKVVN--HVQNNISIHWHGIRQLQSGWADGPAY 107
C ++ +NG+FPGP I A GD L+I + N H + + IHWHGIRQ + WADG A
Sbjct: 168 CKEHVVMGINGQFPGPTIRAEVGDTLVIDLTNKLHTEGTV-IHWHGIRQFGTPWADGTAA 344
Query: 108 VTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRAT-LYGSLII-LPKHNAQYPFVKPY 165
++QC I G+++ Y + + + GT F+H H RA LYGSLI+ LPK + PF
Sbjct: 345 ISQCAINPGETFQYKFKVD-RPGTYFYHGHYGMQRAAGLYGSLIVDLPKSQRE-PFHYDG 518
Query: 166 KEVPMIFGEWFNADPE---AIISQALQTGGGPNVSDAYTINGLPGPLYNCSKKDTFK--- 219
+ ++ W + E + S ++ G P + ING +NCS +
Sbjct: 519 EFNLLLSDLWHTSSHEQEVGLSSAPMRWIGEPQ---SLLINGRGQ--FNCSLASKYGSTN 683
Query: 220 ----------------LKVKPGKIYLLRLINAALNDELFFSIANHTLKVVEADAVYVKPF 263
L V+P K Y +R+ + L +I+NH L VVEAD YV PF
Sbjct: 684 LPQCNLKGGEECAPQILHVEPKKTYRIRIASTTSLASLNLAISNHKLIVVEADGNYVHPF 863
Query: 264 VTKTILITPGQTTNVLLKTKSHYPNATFFMTARPYVTGLGTFDNSTVAG--ILEYKIQHN 321
I I G+T +VLL T P ++++ G+ ST IL YK
Sbjct: 864 AVDDIDIYSGETYSVLLTTDQD-PKKNYWLSI-----GVRGRKPSTPQALTILNYKPL-- 1019
Query: 322 ITHHNSAVKLKKLPLFKPLLPALNDTSFATKFSNKLRSLASARFPANVPQKVDKHFLFTV 381
SA P P+ P NDT + F+ ++ S P + H L T
Sbjct: 1020-----SASVFPTSP--PPVTPLWNDTDHSKAFTKQIISKMGNPQPPKSSHRTI-HLLNT- 1172
Query: 382 GLGTNPCKNKSNQTCQGPNNATMFSGSVNNVSFILPTTALLQAHFFGKSNGVYTPDFPTS 441
+NK + ++NNVS LPTT L + F N P
Sbjct: 1173-------QNKIG---------SFTKWAINNVSLTLPTTPYLGSIKFNLKNTF--DKNPPP 1298
Query: 442 PLHPFNYT-GTTPNNTNVSNGTKVVVLPFNTSVELVMQDTSIL---GAESHPLHLHGFNF 497
P +Y P N N + G V N V++++Q+ + L G+E HP HLHG +F
Sbjct: 1299ERFPMDYDIFKNPVNPNTTTGNGVYTFQLNEVVDVILQNANQLNGNGSEIHPWHLHGHDF 1478
Query: 498 FVVGQGFGNFDPNKDPSNFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHT 557
+V+G G G F P D +FNL RNT + GW A+RF ADNPGVW HCH+E H
Sbjct: 1479WVLGYGEGRFRPGVDERSFNLTRAPLRNTAVIFPYGWTALRFKADNPGVWAFHCHIEPHL 1658
Query: 558 SWGLKMAWVVLDGKLPN 574
G+ + + K+ N
Sbjct: 1659HMGMGVIFAEAVHKVRN 1709
>TC81351 similar to PIR|T00579|T00579 probable laccase [imported] -
Arabidopsis thaliana, partial (27%)
Length = 765
Score = 194 bits (493), Expect = 8e-50
Identities = 86/155 (55%), Positives = 111/155 (71%), Gaps = 1/155 (0%)
Frame = +3
Query: 434 YTPDFPTSPLHPFNYTGTTPNNT-NVSNGTKVVVLPFNTSVELVMQDTSILGAESHPLHL 492
+T DFP FNYTG P GTK+ L + ++V++V+QDTSI+ E HP+H+
Sbjct: 54 FTTDFPPVLPIQFNYTGNVPRGLWTPRKGTKLFKLKYGSNVQIVLQDTSIVTVEDHPMHV 233
Query: 493 HGFNFFVVGQGFGNFDPNKDPSNFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCH 552
HGF+FFVVG GFGNF+P DP+ FNLVDP RNT+G P GGWVAIRF ADNPG+WF+HCH
Sbjct: 234 HGFHFFVVGSGFGNFNPRTDPATFNLVDPPVRNTIGTPPGGWVAIRFKADNPGIWFLHCH 413
Query: 553 LEVHTSWGLKMAWVVLDGKLPNQKLLPPPVDLPKC 587
++ H +WGL A +V +G P Q ++PPP DLP+C
Sbjct: 414 IDSHLNWGLGTALLVENGVGPLQSVIPPPPDLPQC 518
>TC93728 similar to GP|1621467|gb|AAB17194.1| laccase {Liriodendron
tulipifera}, partial (21%)
Length = 514
Score = 193 bits (490), Expect = 2e-49
Identities = 83/113 (73%), Positives = 104/113 (91%)
Frame = +2
Query: 41 IRYQNVTRLCHSKSMVTVNGEFPGPHIVAREGDRLIIKVVNHVQNNISIHWHGIRQLQSG 100
I+ QNVTRLC +KS+VTVNG+FPGP I+AREGDR+++KVVNHVQ N+SIHWHGIRQ++S
Sbjct: 176 IKLQNVTRLCQTKSIVTVNGQFPGPRIIAREGDRVVVKVVNHVQYNVSIHWHGIRQVRSA 355
Query: 101 WADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRATLYGSLIILP 153
WADGPAY+TQCPI+ GQSYV+ +TI GQRGTL++HAHISWLR+TL+G ++ILP
Sbjct: 356 WADGPAYITQCPIKPGQSYVHKFTIIGQRGTLWYHAHISWLRSTLHGPIVILP 514
>AW126324 similar to GP|20161818|db putative laccase {Oryza sativa (japonica
cultivar-group)}, partial (19%)
Length = 535
Score = 189 bits (480), Expect = 3e-48
Identities = 82/112 (73%), Positives = 95/112 (84%)
Frame = +2
Query: 476 VMQDTSILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSNFNLVDPVERNTVGVPSGGWV 535
V+QDT++L ESHP HLHG+NFFVVG G GNFDP KDP+ +NLVDP+ERNTVGVP+GGW
Sbjct: 2 VLQDTNLLTVESHPFHLHGYNFFVVGTGIGNFDPAKDPAKYNLVDPMERNTVGVPTGGWT 181
Query: 536 AIRFLADNPGVWFMHCHLEVHTSWGLKMAWVVLDGKLPNQKLLPPPVDLPKC 587
AIRF A+NPGVWFMHCHLE+HT WGLK A+VV DG +Q +LPPP DLPKC
Sbjct: 182 AIRFTANNPGVWFMHCHLELHTGWGLKTAFVVEDGPGKDQSVLPPPKDLPKC 337
>TC86426 similar to PIR|T07634|T07634 pollen-specific protein homolog
T1P17.10 - Arabidopsis thaliana, partial (91%)
Length = 2220
Score = 187 bits (476), Expect = 7e-48
Identities = 169/596 (28%), Positives = 262/596 (43%), Gaps = 11/596 (1%)
Frame = +3
Query: 3 FLIRLKIYTLLYLFISLCYILFYSFVCSSCNKIRLLTQIRYQNVTRLCHSKSMVTVNGEF 62
FL + I+ LL L + + Y F ++ Y + L + ++ +N +F
Sbjct: 195 FLFLINIFFLLTLSYAEDAFVPYEF------------EVSYITASPLGVPQQVIAINKQF 338
Query: 63 PGPHIVAREGDRLIIKVVNHVQNNISIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYN 122
PGP I + + + V N + N+ IHW G++Q +S W DG T CPI ++ Y
Sbjct: 339 PGPTINVTTNNNVAVNVHNKLDENLLIHWSGVQQRRSSWQDG-VLGTNCPIPPKWNWTYQ 515
Query: 123 YTIKGQRGTLFWHAHISWLRAT-LYGSLIILPKHNAQYPFVKPYKEVPMIFGEWFNADPE 181
+ +K Q G+ F+ +++ RA +G II + PF P +++ + G+W+ +
Sbjct: 516 FQVKDQIGSFFYFPSLNFQRAAGGFGGFIINNRPVISVPFDTPERDIVVFIGDWYTRNHT 695
Query: 182 AIISQALQTGGGPNVSDAYTINGLPGPLYNCSKK----DTFKLKVKPGKIYLLRLINAAL 237
A + +AL G + D ING YN + D ++ VKPGK Y LR+ N +
Sbjct: 696 A-LRKALDDGKDLGMPDGVLINGKGPYRYNDTLVPEGIDFEQIDVKPGKTYRLRVHNVGI 872
Query: 238 NDELFFSIANHTLKVVEADAVYVKPFVTKTILITPGQTTNVLLKTKSHYPNATFFMTARP 297
+ L F I NH L + E + Y ++ I GQ+ + L+ T ++ +++ A
Sbjct: 873 STSLNFRIQNHNLLLAETEGSYTVQQNYTSLDIHVGQSYSFLV-TMDQNASSDYYIVASA 1049
Query: 298 YVTGLGTFDNSTVAGILEYKIQHNITHHNSAVKLKKLPLFKPLLPALNDTSFATKFS-NK 356
+ T GIL Y NS K + LP D F FS N+
Sbjct: 1050RFVNESRWQRVTGVGILHYS--------NSKGKA------RGHLPPGPDDQFDKTFSMNQ 1187
Query: 357 LRSL---ASARFPANVPQKVDKHFLFTVGLGTNPCKNKSNQTCQGPNNATMFSGSVNNVS 413
RS+ SA PQ ++ V KNK G AT+ + +S
Sbjct: 1188ARSIRWNVSASGARPNPQGSFRYGSINV-TEIYVLKNKPPVKIDGKRRATL-----SGIS 1349
Query: 414 FILPTTALLQAHFFGKSNGVYTPDFPTSPLHPFNYTGTTPNNTNVSNGTKVVVLPFNTSV 473
F P T + A + K GVY DFPT PL TG+ T+V NG+ F +
Sbjct: 1350FANPATPIRLADHY-KLKGVYKLDFPTKPL-----TGSPRVETSVINGS------FRGFM 1493
Query: 474 ELVMQDTSILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSNFNLVDPVERNTVGVPSGG 533
E+++Q+ + H HL G+ FFVVG FG++ N +N D + R+T V G
Sbjct: 1494EIILQNND---TKMHTYHLSGYAFFVVGMDFGDWSEN-SRGTYNKWDGIARSTAQVYPGA 1661
Query: 534 WVAIRFLADNPGVWFMHCHLEVHTSW--GLKMAWVVLDGKLPNQKLLPPPVDLPKC 587
W A+ DN GVW + E SW G + V++ + N+ LP P + KC
Sbjct: 1662WTAVLVSLDNVGVW--NLRTENLDSWYLGQETYIRVVNPEPTNKTELPMPDNALKC 1823
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,608,356
Number of Sequences: 36976
Number of extensions: 347727
Number of successful extensions: 2248
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 2153
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2188
length of query: 587
length of database: 9,014,727
effective HSP length: 101
effective length of query: 486
effective length of database: 5,280,151
effective search space: 2566153386
effective search space used: 2566153386
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0342a.1


