

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0336.21
(656 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
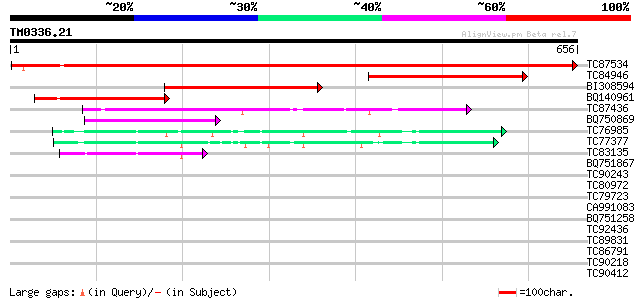
Score E
Sequences producing significant alignments: (bits) Value
TC87534 similar to SP|P17597|ILVB_ARATH Acetolactate synthase c... 1099 0.0
TC84946 similar to GP|16245|emb|CAA35887.1|| precursor acetolact... 320 8e-88
BI308594 similar to GP|1130684|emb acetohydroxyacid synthase {Go... 310 1e-84
BQ140961 similar to SP|P09342|ILV1 Acetolactate synthase I chlo... 217 1e-56
TC87436 similar to PIR|T51575|T51575 2-hydroxyphytanoyl-CoA lyas... 128 8e-30
BQ750869 weakly similar to GP|21205303|db alpha-acetolactate syn... 98 1e-20
TC76985 homologue to GP|17225598|gb|AAL37492.1 pyruvate decarbox... 86 5e-17
TC77377 homologue to SP|P51850|DCP1_PEA Pyruvate decarboxylase i... 85 8e-17
TC83135 similar to GP|10187157|emb|CAC09052. cDNA~Strawberry pyr... 63 3e-10
BQ751867 similar to EGAD|117950|126 hypothetical protein F11E6.f... 34 0.16
TC90243 similar to GP|21166059|gb|AAM43676.1 SWIFT. 6/101 {Dicty... 32 0.77
TC80972 weakly similar to GP|23496972|gb|AAN36523.1 hypothetical... 30 2.3
TC79723 homologue to SP|Q01520|EF1A_PODAN Elongation factor 1-al... 30 2.3
CA991083 similar to GP|15450401|gb At1g09870/F21M12_26 {Arabidop... 30 2.3
BQ751258 weakly similar to GP|6224824|gb|A PHR1 homolog {Pneumoc... 30 2.3
TC92436 similar to GP|6862922|gb|AAF30311.1| unknown protein {Ar... 30 2.9
TC89831 similar to PIR|H96569|H96569 unknown protein 54928-5675... 30 2.9
TC86791 similar to GP|9828617|gb|AAG00240.1| F1N21.15 {Arabidops... 30 2.9
TC90218 homologue to SP|P48490|PP1_PHAVU Serine/threonine protei... 30 3.8
TC90412 homologue to SP|P23704|ATPB_NEUCR ATP synthase beta chai... 29 5.0
>TC87534 similar to SP|P17597|ILVB_ARATH Acetolactate synthase chloroplast
precursor (EC 4.1.3.18) (Acetohydroxy-acid synthase)
(ALS)., partial (87%)
Length = 2258
Score = 1099 bits (2842), Expect = 0.0
Identities = 556/658 (84%), Positives = 599/658 (90%), Gaps = 4/658 (0%)
Frame = +2
Query: 3 AAPAAKLAFTTV---PSPSSRP-QNHVLKFTIPFPSYPNNSSPHSRCRSLQISASLSNPT 58
AA AAK AFTT SP S P QNHV + T PF +YPN + RSL+ISAS+SNPT
Sbjct: 59 AATAAKAAFTTTISSSSPQSLPNQNHVFRITYPFSTYPNTPFKLNH-RSLKISASVSNPT 235
Query: 59 PTAASASTAVATENFTSRFSLDEPRKGADILVEALERQGVTDVFAYPGGASMEIHQALTR 118
+ + E F SRF+ +EPRKGADILVE+LERQGVT+VFAYPGGASMEIHQALTR
Sbjct: 236 Q---KTTPIPSPEQFISRFAPNEPRKGADILVESLERQGVTNVFAYPGGASMEIHQALTR 406
Query: 119 SATIRNVLPRHEQGGIFAAEGYARSSGLPGVCIATSGPGATNLVSGLADALLDSVPLIAI 178
S IRNVLPRHEQGGIFAAEGYARSSGLPGVCIATSGPGATNLVSGLADA+LDSVPLIAI
Sbjct: 407 STAIRNVLPRHEQGGIFAAEGYARSSGLPGVCIATSGPGATNLVSGLADAMLDSVPLIAI 586
Query: 179 TGQVPRRMIGTDAFQETPIVEVTRSITKHNYLVLDIDDIPRIVNEAFFLATSGRPGPVLI 238
TGQVPRRMIGTDAFQETPIVEVTRSITKHNYLVLD+DDIPRIV EAF LA+SGRPGPVLI
Sbjct: 587 TGQVPRRMIGTDAFQETPIVEVTRSITKHNYLVLDVDDIPRIVKEAFLLASSGRPGPVLI 766
Query: 239 DIPKDIQQQLAVPNWDQPIKLPGYMARLPRSPSDALLEQVVRLLLECKKPVLYAGGGSLN 298
DIPKDIQQQ+++PNWDQPI+L GYM RLP++P +A LEQ+VRLLLE KKPVLY GGGSLN
Sbjct: 767 DIPKDIQQQVSLPNWDQPIRLTGYMNRLPKAPDEAHLEQIVRLLLESKKPVLYVGGGSLN 946
Query: 299 SSEELRRFVELTGVPVASTLMGLGAYPTADENSLQMLGMHGTVYANYAVDKSDLLLAFGV 358
SEELRRFVELTGVPVASTLMGLG+YP+ DENSLQMLGMHGTVYANYAVDKSDLLLAFGV
Sbjct: 947 CSEELRRFVELTGVPVASTLMGLGSYPSLDENSLQMLGMHGTVYANYAVDKSDLLLAFGV 1126
Query: 359 RFDDRVTGKLEAFASRAKIVHIDIDSAEIGKNKQPHVSVCADLKLALKGINRILESRGVK 418
RFDDRVTGKLEAFASRAKIVHIDID+AEIGKNKQPHVSVC DLKLALKGIN+ILES G++
Sbjct: 1127RFDDRVTGKLEAFASRAKIVHIDIDAAEIGKNKQPHVSVCGDLKLALKGINQILESNGIE 1306
Query: 419 GKLDFRAWREELNEQKLRFPLQFKTFEEAIPPQYAIQVLDELTNGNAIISTGVGQHQMWA 478
KLDF WREELN+QK+RFP+ +KTF+EAIPPQYAIQVLDELTNG AIISTGVGQHQMWA
Sbjct: 1307RKLDFGGWREELNDQKVRFPMSYKTFDEAIPPQYAIQVLDELTNGEAIISTGVGQHQMWA 1486
Query: 479 AQFYKYKRPRQWLTSSGLGAMGFGLPAAMGAAVANPGDVVVDIDGDGSFMMNVQELATIK 538
AQFY+YKRPRQ LTS+GLGAMGFGLPAAMGAAVANP +VVDIDGDGSFMMNVQELATIK
Sbjct: 1487AQFYRYKRPRQRLTSAGLGAMGFGLPAAMGAAVANPDAIVVDIDGDGSFMMNVQELATIK 1666
Query: 539 VENLPVKILLLNNQHLGMVVQWEDRFYKSNRAHTYLGDPSNEKEVFPNMLKFADACGIPA 598
VENLPVKILLLNNQHLGMVVQWEDRFYK+NRAHTYLGDP+NEKE+FPNMLKFA ACGIPA
Sbjct: 1667VENLPVKILLLNNQHLGMVVQWEDRFYKANRAHTYLGDPANEKEIFPNMLKFAGACGIPA 1846
Query: 599 ARVTKKQDLRAAIQKMLDTPGPYLLDVIVPHQEHVLPMIPSNGTFQDVITEGDGRRSY 656
ARVTK+ DLRAAIQKMLDTPGPYLLDVIVPHQEHVLPMIP+NG+F+DVITEGDGR SY
Sbjct: 1847ARVTKRADLRAAIQKMLDTPGPYLLDVIVPHQEHVLPMIPANGSFEDVITEGDGRISY 2020
>TC84946 similar to GP|16245|emb|CAA35887.1|| precursor acetolactate
synthase (670 AA) {Arabidopsis thaliana}, partial (26%)
Length = 558
Score = 320 bits (821), Expect = 8e-88
Identities = 157/185 (84%), Positives = 169/185 (90%), Gaps = 1/185 (0%)
Frame = +3
Query: 416 GVKGKLDFRAWREELNEQKLRFPLQFKTFEEAIPPQYAIQVLDELTNGNAIISTGVGQHQ 475
G+KGKLDF AWR+ELN QKL+FPL FKTFE+AI PQYAIQVLDELTNG+AI+STGVGQHQ
Sbjct: 3 GIKGKLDFEAWRQELNVQKLKFPLGFKTFEDAISPQYAIQVLDELTNGDAIVSTGVGQHQ 182
Query: 476 MWAAQFYKYKRPRQWLTSSGLGAMGFGLPAAMGAAVANPGDVVVDIDGDGSFMMNVQELA 535
MW+AQFYKYKRPRQWLTS GLGAMGFGLPAA+GAAVANP +VVDIDGDGSFMMNVQELA
Sbjct: 183 MWSAQFYKYKRPRQWLTSGGLGAMGFGLPAAIGAAVANPDAIVVDIDGDGSFMMNVQELA 362
Query: 536 TIKVENLPVKILLLNNQHLGMVVQWEDRFYKSNRAHTYLGDPSNEKEVFP-NMLKFADAC 594
TI+VENLPVKILLLNNQHLGMVVQWEDRFYK+NRAHTYLGDP FP + L FADAC
Sbjct: 363 TIRVENLPVKILLLNNQHLGMVVQWEDRFYKANRAHTYLGDPF*XG*DFP*HCLDFADAC 542
Query: 595 GIPAA 599
GIPAA
Sbjct: 543 GIPAA 557
>BI308594 similar to GP|1130684|emb acetohydroxyacid synthase {Gossypium
hirsutum}, partial (27%)
Length = 554
Score = 310 bits (794), Expect = 1e-84
Identities = 149/182 (81%), Positives = 167/182 (90%)
Frame = +2
Query: 180 GQVPRRMIGTDAFQETPIVEVTRSITKHNYLVLDIDDIPRIVNEAFFLATSGRPGPVLID 239
GQVPRRMIGTDAFQETPIVEVTRSITKHNYL+LD++DIPR+V EAFFLATSGRPGPVLID
Sbjct: 2 GQVPRRMIGTDAFQETPIVEVTRSITKHNYLILDVEDIPRVVKEAFFLATSGRPGPVLID 181
Query: 240 IPKDIQQQLAVPNWDQPIKLPGYMARLPRSPSDALLEQVVRLLLECKKPVLYAGGGSLNS 299
+PKD+QQQLAVPNW +PIKL GY++RLP+ P +A LEQV+RLLLE +KPVLY GGG LNS
Sbjct: 182 VPKDVQQQLAVPNWSEPIKLTGYLSRLPKIPGEAQLEQVLRLLLESEKPVLYVGGGCLNS 361
Query: 300 SEELRRFVELTGVPVASTLMGLGAYPTADENSLQMLGMHGTVYANYAVDKSDLLLAFGVR 359
S+EL+RFVELTGVPVASTLMGLG+YP E+SL MLGMHGTVYANYAVD SDLLLAFGV+
Sbjct: 362 SDELKRFVELTGVPVASTLMGLGSYPIGGEHSLSMLGMHGTVYANYAVDNSDLLLAFGVK 541
Query: 360 FD 361
D
Sbjct: 542 VD 547
>BQ140961 similar to SP|P09342|ILV1 Acetolactate synthase I chloroplast
precursor (EC 4.1.3.18) (Acetohydroxy-acid synthase I)
(ALS I)., partial (17%)
Length = 466
Score = 217 bits (553), Expect = 1e-56
Identities = 114/156 (73%), Positives = 127/156 (81%)
Frame = +1
Query: 29 TIPFPSYPNNSSPHSRCRSLQISASLSNPTPTAASASTAVATENFTSRFSLDEPRKGADI 88
T+ FP P + P + + IS SL TA ++T E FTSRFS +PRKG+DI
Sbjct: 4 TLTFPLSPILNKPKTTHSLIGISCSLK--PFTAPPSTTTTVDEPFTSRFSSTQPRKGSDI 177
Query: 89 LVEALERQGVTDVFAYPGGASMEIHQALTRSATIRNVLPRHEQGGIFAAEGYARSSGLPG 148
LVEALER+GVT+VFAYPGGASMEIHQALTRS TIRN+LPRHEQGG+FAAEGYARSSGLPG
Sbjct: 178 LVEALEREGVTNVFAYPGGASMEIHQALTRSKTIRNILPRHEQGGVFAAEGYARSSGLPG 357
Query: 149 VCIATSGPGATNLVSGLADALLDSVPLIAITGQVPR 184
VCIATSGPGATNL SGLADAL+DSVPLIAITGQVPR
Sbjct: 358 VCIATSGPGATNLXSGLADALMDSVPLIAITGQVPR 465
>TC87436 similar to PIR|T51575|T51575 2-hydroxyphytanoyl-CoA lyase-like
protein - Arabidopsis thaliana, partial (75%)
Length = 1592
Score = 128 bits (321), Expect = 8e-30
Identities = 112/474 (23%), Positives = 209/474 (43%), Gaps = 24/474 (5%)
Frame = +1
Query: 85 GADILVEALERQGVTDVFAYPGGASMEIHQALTRSAT--IRNVLPRHEQGGIFAAEGYAR 142
G + ++ + GV +F G + + TR+ + IR + +EQ +AA Y
Sbjct: 193 GNVLAAKSFSQFGVLHMFGVVG---IPVTSLATRAVSLGIRFIAFHNEQSAGYAASAYGY 363
Query: 143 SSGLPGVCIATSGPGATNLVSGLADALLDSVPLIAITGQVPRRMIGTDAFQETPIVEVTR 202
+ PG+ + SGPG + ++GL++ ++ P + I+G + +G FQE ++ +
Sbjct: 364 LTSRPGIFLTVSGPGCVHGLAGLSNGTTNTWPTVMISGSCNQNDVGRGDFQELDQIQAVK 543
Query: 203 SITKHNYLVLDIDDIPRIVNEAFFLATSGRPGPVLIDIPKDI-QQQLAVPNWDQPIKLPG 261
TK I +IP V + + RPG V +D+P D+ Q+++ + +
Sbjct: 544 PFTKFAIKAKHISEIPNCVAQVLANSVVNRPGGVYLDLPTDVLHQKVSESEAESLLTEAK 723
Query: 262 YMARLPR-----SPSDALLEQVVRLLLECKKPVLYAGGGS--LNSSEELRRFVELTGVPV 314
Y+A + S + +++ V LL + ++P++ G G+ + +EL++ VE TG+P
Sbjct: 724 YLAEKIKKSHVISVESSKIQEAVSLLRKAERPLIVFGKGAAYAKAEDELKKLVEKTGIPF 903
Query: 315 ASTLMGLGAYPTADENSLQMLGMHGTVYANYAVDKSDLLLAFGVRFDDRV-TGKLEAFAS 373
T MG G P D++ L + + A+ K D+ + G R + + G+ ++
Sbjct: 904 LPTPMGKGLLP--DDHPLA-----ASAARSLAIGKCDVAIVIGARLNWLLHFGEEPKWSK 1062
Query: 374 RAKIVHIDIDSAEIGKNKQPHVSVCADLKLALKGINRILES------------RGVKGKL 421
K V +D+ EI + ++P + + D K L+ +N+ ++ + K+
Sbjct: 1063DVKFVLVDVSKEEI-ELRKPFLGLVGDGKEVLEVLNKEIKDDPFCLGSTHPWVEAISSKV 1239
Query: 422 DFRAWREELNEQKLRFPLQFKTFEEAIPPQYAIQVLDELTNGNAIISTGVGQHQMWAAQ- 480
+ E K P F T P + + E A + G + M +
Sbjct: 1240KDNGVKMEAQLAKDVVPFNFLT-----PMRIIRDAISEFGGSPAPVVVSEGANTMDVGRS 1404
Query: 481 FYKYKRPRQWLTSSGLGAMGFGLPAAMGAAVANPGDVVVDIDGDGSFMMNVQEL 534
K PR L + G MG GL + AAVA P +VV ++GD + + E+
Sbjct: 1405VLVQKEPRTRLDAGTWGTMGVGLGYCIAAAVAYPDRLVVAVEGDSGYGFSAMEV 1566
>BQ750869 weakly similar to GP|21205303|db alpha-acetolactate synthase
{Staphylococcus aureus subsp. aureus MW2}, partial (18%)
Length = 732
Score = 97.8 bits (242), Expect = 1e-20
Identities = 55/158 (34%), Positives = 81/158 (50%)
Frame = +1
Query: 87 DILVEALERQGVTDVFAYPGGASMEIHQALTRSATIRNVLPRHEQGGIFAAEGYARSSGL 146
D+++++L GV +F PG + AL I+ V+ RHEQ F A R +G
Sbjct: 70 DVVIDSLIAAGVKHIFGVPGAKIDSVFNALIDRPEIKLVVCRHEQNAAFIAGAIGRITGR 249
Query: 147 PGVCIATSGPGATNLVSGLADALLDSVPLIAITGQVPRRMIGTDAFQETPIVEVTRSITK 206
PGVCIATSGPG +NLV+GL A + P++A+ G V R Q V++ +TK
Sbjct: 250 PGVCIATSGPGTSNLVTGLVTANDEGAPVVALVGDVKRVQAAKKTHQSLRGVQLLEPVTK 429
Query: 207 HNYLVLDIDDIPRIVNEAFFLATSGRPGPVLIDIPKDI 244
+ D I ++ +AF AT+ G + +P DI
Sbjct: 430 KATGAVHPDQIAELMLDAFRTATAHPQGATAVSLPIDI 543
>TC76985 homologue to GP|17225598|gb|AAL37492.1 pyruvate decarboxylase
{Fragaria x ananassa}, partial (94%)
Length = 2130
Score = 85.9 bits (211), Expect = 5e-17
Identities = 125/552 (22%), Positives = 217/552 (38%), Gaps = 26/552 (4%)
Frame = +1
Query: 50 ISASLSNPTPTAASASTAVATENFTSRFSLDEPRKGADILVEALERQGVTDVFAYPGGAS 109
+S N T T STA+AT + T L L + GVTDVF+ PG +
Sbjct: 175 VSCKQQNSTATI-QPSTAIATSDATL----------GRHLARRLVQVGVTDVFSVPGDFN 321
Query: 110 MEIHQALTRSATIRNVLPRHEQGGIFAAEGYARSSGLPGVCIATSGPGATNLVSGLADAL 169
+ + L + + +E +AA+GYARS G+ G C+ T G ++++ +A A
Sbjct: 322 LTLLDHLINEPELNLIGCCNELNAGYAADGYARSRGV-GACVVTFTVGGLSVLNAIAGAY 498
Query: 170 LDSVPLIAITG----------QVPRRMIGTDAFQETPIVEVTRSITKHNYLVLDIDDIPR 219
+++PLI I G ++ IG F + + +++T +V +++D
Sbjct: 499 SENLPLICIVGGPNSNDYGTSRILHHTIGVPDFSQE--LRCFQTVTCFQAVVNNLEDAHE 672
Query: 220 IVNEAFFLA-TSGRP-----GPVLIDIPKDIQQQLAVPNWDQPIKLPGYMARLPRSPSDA 273
+++ A A +P G L IP + VP P KL +M +A
Sbjct: 673 LIDTAISTALKESKPVYISIGCNLPGIPHPTFSRDPVPFSLAP-KLSNHMG------LEA 831
Query: 274 LLEQVVRLLLECKKPVLYAGGGSLNSSEELRRFVELTGVPVASTLMGLGAYPTADENSLQ 333
+E L + KPVL GG L ++ FVEL + + A E+
Sbjct: 832 AVEAAAEFLNKAVKPVL-VGGPKLRVAKASDAFVELADASGYALAVMPSAKGMVPEHHPH 1008
Query: 334 MLGMH----GTVYANYAVDKSDLLLAFGVRFDDRVTGKLEAFASRAKIVHIDIDSAEIGK 389
+G + T + V+ +D L G F+D + + K + + D +
Sbjct: 1009FIGTYWGAVSTAFCAEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQPDRVVVAN 1188
Query: 390 NKQPHVSVCADLKLALKGINRILESRGVKGKLDFRAW------REELNEQKLRFPLQFKT 443
C +K LK + + L+ + R + + ++ LR + F+
Sbjct: 1189GP---AFGCILMKDFLKALAKRLKHNNAAYENYHRIFVPDGKPLKSAPKEPLRVNVMFQH 1359
Query: 444 FEEAIPPQYAIQVLDELTNGNAIISTGVGQHQMWAAQFYKYKRPRQWLTSSGLGAMGFGL 503
++ + + A+ I TG + Q K + G++G+ +
Sbjct: 1360IQQMLSRETAV-----------IAETG---DSWFNCQKLKLPEGCGYEFQMQYGSIGWSV 1497
Query: 504 PAAMGAAVANPGDVVVDIDGDGSFMMNVQELATIKVENLPVKILLLNNQHLGMVVQWEDR 563
A +G A A P V+ GDGSF + Q+++T+ I L+NN + V+ D
Sbjct: 1498GATLGYAQAVPEKRVIACIGDGSFQVTAQDVSTMLRCGQKTIIFLINNGGYTIEVEIHDG 1677
Query: 564 FYKSNRAHTYLG 575
Y + Y G
Sbjct: 1678PYNVIKNWNYTG 1713
>TC77377 homologue to SP|P51850|DCP1_PEA Pyruvate decarboxylase isozyme 1
(EC 4.1.1.1) (PDC). [Garden pea] {Pisum sativum},
partial (95%)
Length = 2128
Score = 85.1 bits (209), Expect = 8e-17
Identities = 125/545 (22%), Positives = 215/545 (38%), Gaps = 30/545 (5%)
Frame = +2
Query: 51 SASLSNPTPTAASASTAVATENFTSRFSLDEPRKGADILVEALERQGVTDVFAYPGGASM 110
S LSN T+ +S FT ++ L L GV DVF+ PG ++
Sbjct: 131 SYKLSNSMETSPPSSAPSTVRPFTCDGTM------GGHLARRLVEIGVRDVFSVPGDFNL 292
Query: 111 EIHQALTRSATIRNVLPRHEQGGIFAAEGYARSSGLPGVCIATSGPGATNLVSGLADALL 170
+ L + V +E +AA+GYAR+ G+ G C+ T G ++++ +A A
Sbjct: 293 TLLDHLIAEPELNLVGCCNELNAGYAADGYARAKGV-GACVVTFTVGGLSILNAIAGAYS 469
Query: 171 DSVPLIAITGQVPRRMIGTDAFQETPI--------VEVTRSITKHNYLVLDIDDIPRIVN 222
+++P+I I G GT+ I + ++IT +V +++D +++
Sbjct: 470 ENLPVICIVGGPNSNDYGTNRILHHTIGLPDFSQELRCFQTITCFQAVVNNLEDAHELID 649
Query: 223 EAFFLATSGRPGPVLIDIPKDIQQQLAVPNWDQPIKLPGYMARLPRSPS----DALLEQV 278
A A PV I I ++ + P + + +P ++A PR + +A +E+
Sbjct: 650 TAISTALK-ESKPVYISIGCNL-PAIPHPTFARD-PVPFFLA--PRVSNQEGLEAAVEEA 814
Query: 279 VRLLLECKKPVLYAGGGSL---NSSEELRRFVELTGVPVASTLMGLGAYPTADENSLQML 335
L + KPV+ GG L + + F E +G P+A G G P EN +
Sbjct: 815 AAFLNKAVKPVI-VGGPKLRVAKAQKAFMEFAEASGYPIAVMPSGKGLVP---ENHPHFI 982
Query: 336 GMH----GTVYANYAVDKSDLLLAFGVRFDDRVTGKLEAFASRAKIVHIDIDSAEIGKNK 391
G + T Y V+ +D + G F+D + + K + + + IG
Sbjct: 983 GTYWGAVSTSYCGEIVESADAYVFVGPIFNDYSSVGYSLLIKKEKSLIVQPNRVTIGNGL 1162
Query: 392 QPHVSVCADLKLAL-----------KGINRILESRGVKGKLDFRAWREELNEQKLRFPLQ 440
AD AL + RI G+ K W + ++ LR +
Sbjct: 1163SLGWVFMADFLTALSKKVKKNTAAVENYRRIYVPPGIPLK-----WEK---DEPLRVNVL 1318
Query: 441 FKTFEEAIPPQYAIQVLDELTNGNAIISTGVGQHQMWAAQFYKYKRPRQWLTSSGLGAMG 500
FK +E + A+ I TG + Q + + G++G
Sbjct: 1319FKHIQELLSGDTAV-----------IAETG---DSWFNCQKLRLPENCGYEFQMQYGSIG 1456
Query: 501 FGLPAAMGAAVANPGDVVVDIDGDGSFMMNVQELATIKVENLPVKILLLNNQHLGMVVQW 560
+ + A +G A A V+ GDGSF + Q+++T+ I L+NN + V+
Sbjct: 1457WSVGATLGYAQAATNKRVIACIGDGSFQVTAQDISTMIRCGQKSIIFLINNGGYTIEVEI 1636
Query: 561 EDRFY 565
D Y
Sbjct: 1637HDGPY 1651
>TC83135 similar to GP|10187157|emb|CAC09052. cDNA~Strawberry pyruvate
decarboxylase {Fragaria x ananassa}, partial (30%)
Length = 700
Score = 63.2 bits (152), Expect = 3e-10
Identities = 43/179 (24%), Positives = 83/179 (46%), Gaps = 8/179 (4%)
Frame = +1
Query: 58 TPTAASASTAVATENFTSRFSLDEPRKGADILVEALERQGVTDVFAYPGGASMEIHQALT 117
TPT+ + + T + E G+ L L G+TD+F PG ++ + L
Sbjct: 109 TPTSNGTVSTIQKSPSTQSLASSESTLGSH-LARRLVEVGITDIFTVPGDFNLTLLDHLI 285
Query: 118 RSATIRNVLPRHEQGGIFAAEGYARSSGLPGVCIATSGPGATNLVSGLADALLDSVPLIA 177
++N+ +E +AA+GYARS G+ G C+ T G ++++ +A A +++P+I
Sbjct: 286 AEPKLKNIGCCNELNAGYAADGYARSRGV-GACVVTFTVGGVSVINAIAGAYSENLPVIC 462
Query: 178 ITGQVPRRMIGTDAFQETPI--------VEVTRSITKHNYLVLDIDDIPRIVNEAFFLA 228
I G GT+ I + +++T + +V +++D +++ A A
Sbjct: 463 IVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQAVVNNLEDAHEMIDTAISTA 639
>BQ751867 similar to EGAD|117950|126 hypothetical protein F11E6.f
{Caenorhabditis elegans}, partial (4%)
Length = 585
Score = 34.3 bits (77), Expect = 0.16
Identities = 24/70 (34%), Positives = 38/70 (54%), Gaps = 2/70 (2%)
Frame = +3
Query: 2 AAAPAAKLAFTTVPSPSSRPQNHVLKFTIPFPSYPNNSSPHSRCRSLQISA--SLSNPTP 59
++AP+A++ F + +RP+ ++P P P +SS SR R +A S PTP
Sbjct: 51 SSAPSARV-FAHTSAARTRPRTAPKACSLPLPDPPTSSSNSSRTRRSGPTA*RP*SEPTP 227
Query: 60 TAASASTAVA 69
A+SAS+ A
Sbjct: 228 PASSASSTPA 257
>TC90243 similar to GP|21166059|gb|AAM43676.1 SWIFT. 6/101 {Dictyostelium
discoideum}, partial (2%)
Length = 775
Score = 32.0 bits (71), Expect = 0.77
Identities = 27/93 (29%), Positives = 39/93 (41%), Gaps = 1/93 (1%)
Frame = +1
Query: 12 TTVPSPSSRPQNHVLKFTIPFPSYPNNSSPHSRCRSLQISASLSNPTPTAASASTAVATE 71
T + S S NH +F F S P+ HS+ + L S SLS P+ +++S+ST
Sbjct: 157 TMLHSTSLLHHNHH-RFLFSFRSKPSLLDSHSQSQPLSFSKSLSLPSSSSSSSSTCCHVA 333
Query: 72 NF-TSRFSLDEPRKGADILVEALERQGVTDVFA 103
T L P G + E + D A
Sbjct: 334 RISTEPLELSTPPPGFNFRREITRLTSLRDKLA 432
>TC80972 weakly similar to GP|23496972|gb|AAN36523.1 hypothetical protein
{Plasmodium falciparum 3D7}, partial (9%)
Length = 662
Score = 30.4 bits (67), Expect = 2.3
Identities = 17/46 (36%), Positives = 28/46 (59%)
Frame = +3
Query: 34 SYPNNSSPHSRCRSLQISASLSNPTPTAASASTAVATENFTSRFSL 79
+YP NSS S S S+S S+ + +A+S+S++ ++F RF L
Sbjct: 525 TYPYNSSCSSSSTSSSSSSSFSSLSSSASSSSSSSIHQDFFDRFIL 662
>TC79723 homologue to SP|Q01520|EF1A_PODAN Elongation factor 1-alpha
(EF-1-alpha). {Podospora anserina}, complete
Length = 1537
Score = 30.4 bits (67), Expect = 2.3
Identities = 18/45 (40%), Positives = 26/45 (57%)
Frame = +3
Query: 29 TIPFPSYPNNSSPHSRCRSLQISASLSNPTPTAASASTAVATENF 73
T P+ P SSP S RS + ASLS TP+++S T +++ F
Sbjct: 1158 TATLPTLPA-SSPRSSRRSTAVPASLSRTTPSSSSLVTPLSSRWF 1289
>CA991083 similar to GP|15450401|gb At1g09870/F21M12_26 {Arabidopsis
thaliana}, partial (34%)
Length = 714
Score = 30.4 bits (67), Expect = 2.3
Identities = 24/76 (31%), Positives = 32/76 (41%), Gaps = 3/76 (3%)
Frame = -1
Query: 82 PRKGADILVEALERQGVTDVFAYPGGASMEIHQALTRSATIRNVLPRHEQGGIFAA---E 138
P KGA +L G + G + L+ S + RN RHE G +A
Sbjct: 330 PAKGATVLPRHFRFCG-----GFGGSSKGFSFWILSNSDSSRNKPSRHENGTTVSA*ANR 166
Query: 139 GYARSSGLPGVCIATS 154
A S GLPG C ++S
Sbjct: 165 SLAFSKGLPGTCFSSS 118
>BQ751258 weakly similar to GP|6224824|gb|A PHR1 homolog {Pneumocystis
carinii}, partial (9%)
Length = 503
Score = 30.4 bits (67), Expect = 2.3
Identities = 19/58 (32%), Positives = 27/58 (45%), Gaps = 4/58 (6%)
Frame = +1
Query: 12 TTVPSPSSRPQNHVLKFTIPFPSYPNNSSPHSRCR----SLQISASLSNPTPTAASAS 65
TT+PSPSS P T+P P + R SL +S++ S PT ++S
Sbjct: 151 TTIPSPSSLPSTAATSPTVPRAVSSRRPPPSTPRR*PTSSLAVSSTCSTRRPTTTASS 324
>TC92436 similar to GP|6862922|gb|AAF30311.1| unknown protein {Arabidopsis
thaliana}, partial (56%)
Length = 710
Score = 30.0 bits (66), Expect = 2.9
Identities = 19/75 (25%), Positives = 34/75 (45%)
Frame = +1
Query: 3 AAPAAKLAFTTVPSPSSRPQNHVLKFTIPFPSYPNNSSPHSRCRSLQISASLSNPTPTAA 62
A A + A +P P + F P S+ +S S RSL S +P+ T++
Sbjct: 67 ATIATRSATVNMPHPPPSTSSSSSSFYSPARSFSPPTSSTSSTRSLSFSLPFISPSLTSS 246
Query: 63 SASTAVATENFTSRF 77
+S++ + + SR+
Sbjct: 247 HSSSSPSVSSSVSRY 291
>TC89831 similar to PIR|H96569|H96569 unknown protein 54928-56750
[imported] - Arabidopsis thaliana, partial (39%)
Length = 897
Score = 30.0 bits (66), Expect = 2.9
Identities = 45/191 (23%), Positives = 78/191 (40%), Gaps = 6/191 (3%)
Frame = +1
Query: 29 TIPFPSY-PNNSSPHSRCRSLQISASLSNPTPTAASASTAVATENFTSRFSLDEPRKGAD 87
T+P ++ P+ S P S+ +S+ P AS NFT R + +
Sbjct: 133 TVPHHTFLPSISKPKSKSKSIS--------KPQNFPASRFSTNPNFTKRAVVATKVSTEE 288
Query: 88 ILVEALERQGVTDVFAYPGGASMEIH----QALTRSATIRNVLPRHEQGGIFAAEGYARS 143
V A++ +TD + +E+ + T T N +E G+ + EG+ S
Sbjct: 289 FDVIAVQSNDITDQQEGLIVSRVEMEGGDGELGTTMTTQVNGFGANE--GLLSLEGFPSS 462
Query: 144 SGLPGVCIATSGPGATNLVSGLADALLDSVPLIAITGQVPRRM-IGTDAFQETPIVEVTR 202
SGL G N+ L ++ S+ L+A T + + + I +D + I E+ R
Sbjct: 463 SGLVG------NENEENVEKLLDRSINASIVLVAGTFALTKLLTIDSDYWHGWTIYEIVR 624
Query: 203 SITKHNYLVLD 213
HN+L +
Sbjct: 625 YAPLHNWLAYE 657
>TC86791 similar to GP|9828617|gb|AAG00240.1| F1N21.15 {Arabidopsis
thaliana}, partial (70%)
Length = 1157
Score = 30.0 bits (66), Expect = 2.9
Identities = 26/70 (37%), Positives = 29/70 (41%), Gaps = 2/70 (2%)
Frame = -2
Query: 15 PSPSSRPQNHVLKFTIPFPSYP--NNSSPHSRCRSLQISASLSNPTPTAASASTAVATEN 72
PSPS LK + PF P NN S H SLSNPTP S A +
Sbjct: 787 PSPS-------LKISPPFSPAPPRNNLSAHR---------SLSNPTPFRKSHQEDYAKQA 656
Query: 73 FTSRFSLDEP 82
+ RF L P
Sbjct: 655 YIVRFLLASP 626
>TC90218 homologue to SP|P48490|PP1_PHAVU Serine/threonine protein
phosphatase PP1 (EC 3.1.3.16). [Kidney bean French
bean], partial (95%)
Length = 1024
Score = 29.6 bits (65), Expect = 3.8
Identities = 23/84 (27%), Positives = 37/84 (43%)
Frame = +2
Query: 281 LLLECKKPVLYAGGGSLNSSEELRRFVELTGVPVASTLMGLGAYPTADENSLQMLGMHGT 340
+LL+ + P+ G ++L R E G P A+ + LG Y + SL+ +
Sbjct: 242 ILLDLRAPIRICGDIH-GQYQDLLRLFEYGGYPPAANYLFLGDYVDRGKQSLETIC---- 406
Query: 341 VYANYAVDKSDLLLAFGVRFDDRV 364
LLLA+ +R+ DRV
Sbjct: 407 -----------LLLAYKIRYPDRV 445
>TC90412 homologue to SP|P23704|ATPB_NEUCR ATP synthase beta chain
mitochondrial precursor (EC 3.6.3.14). {Neurospora
crassa}, partial (67%)
Length = 1181
Score = 29.3 bits (64), Expect = 5.0
Identities = 26/98 (26%), Positives = 43/98 (43%), Gaps = 4/98 (4%)
Frame = +3
Query: 2 AAAPAAKLAFTTVPSPS---SRPQNHVLKFTIPFPSYPNNSSPHSRCRSLQISASLSNPT 58
A P TT P P+ S + + ++PF + N S S+ + +S+S P+
Sbjct: 699 AGPPYLSARTTTTPPPACSRSSRSTSLSRISLPFWVWTNFLRLTSSLLSVPVRSSVSCPS 878
Query: 59 PTAASASTAVA-TENFTSRFSLDEPRKGADILVEALER 95
P+ + S+ V+ + TSR L R + V A R
Sbjct: 879 PSPSPRSSLVSRVSSLTSRTPLLLSRPFCPVRVTAFPR 992
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,109,251
Number of Sequences: 36976
Number of extensions: 222426
Number of successful extensions: 1369
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 1348
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1361
length of query: 656
length of database: 9,014,727
effective HSP length: 102
effective length of query: 554
effective length of database: 5,243,175
effective search space: 2904718950
effective search space used: 2904718950
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0336.21


