

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0336.10
(114 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
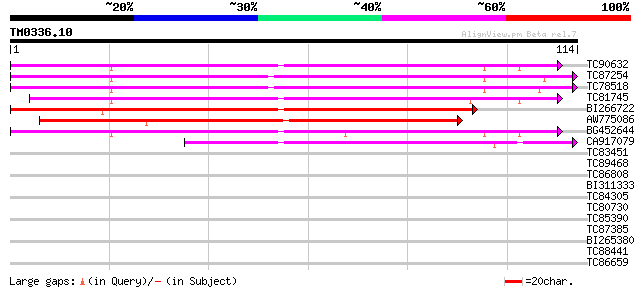
Score E
Sequences producing significant alignments: (bits) Value
TC90632 similar to GP|21554053|gb|AAM63134.1 putative VAMP-assoc... 82 2e-17
TC87254 similar to PIR|JC7234|JC7234 27k vesicle-associated memb... 80 9e-17
TC78518 similar to PIR|JC7234|JC7234 27k vesicle-associated memb... 78 4e-16
TC81745 similar to PIR|JC7234|JC7234 27k vesicle-associated memb... 77 8e-16
BI266722 similar to GP|17473763|gb| putative protein {Arabidopsi... 76 2e-15
AW775086 similar to PIR|JC7234|JC7 27k vesicle-associated membra... 68 5e-13
BG452644 similar to GP|21554053|gb putative VAMP-associated prot... 68 6e-13
CA917079 weakly similar to PIR|B86220|B86 protein F22O13.31 [imp... 52 4e-08
TC83451 weakly similar to GP|19310653|gb|AAL85057.1 unknown prot... 28 0.53
TC89468 similar to GP|5305145|emb|CAB46084.1 fructose-1 6-bispho... 27 1.5
TC86808 homologue to SP|Q9ZTR1|SPD1_PEA Spermidine synthase 1 (E... 26 2.0
BI311333 25 3.4
TC84305 weakly similar to PIR|T39881|T39881 preg-like protein - ... 25 3.4
TC80730 weakly similar to GP|20198248|gb|AAD26886.2 expressed pr... 25 3.4
TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 ... 25 4.4
TC87385 similar to GP|23093788|gb|AAF50220.2 CG16711-PA {Drosoph... 25 5.8
BI265380 weakly similar to GP|17932898|em isoamylase {Triticum a... 24 7.6
TC88441 similar to GP|14194139|gb|AAK56264.1 AT3g06960/F17A9_11 ... 24 7.6
TC86659 similar to GP|5360263|dbj|BAA81904.1 NtPRp27 {Nicotiana ... 24 7.6
>TC90632 similar to GP|21554053|gb|AAM63134.1 putative VAMP-associated
protein {Arabidopsis thaliana}, partial (63%)
Length = 681
Score = 82.4 bits (202), Expect = 2e-17
Identities = 46/119 (38%), Positives = 72/119 (59%), Gaps = 8/119 (6%)
Frame = +1
Query: 1 MDHAEILQIEPAELTFLTE--KKDFCKLHLINKTTNYVAFRLRLSKPEKYGVTPREGGII 58
M ++L IEP EL FL E K+ C L L NKT +YVAF+++ + P KY V P G++
Sbjct: 169 MSTGDLLNIEPVELKFLFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNT-GVV 345
Query: 59 MPQGTYDIIVSRKSDKLPPPGFECTDRFFIQSMALS----VKDLFTE--NRGKRHVEEK 111
+PQ T ++IV+ ++ K PP +C D+F +QS+ ++ K++ E N+ HV E+
Sbjct: 346 LPQSTCEVIVTMQAQKEVPPDMQCKDKFLLQSVRVNDGADAKEITPEMFNKEAGHVVEE 522
>TC87254 similar to PIR|JC7234|JC7234 27k vesicle-associated membrane
protein-associated protein - curled-leaved tobacco,
partial (87%)
Length = 1040
Score = 80.5 bits (197), Expect = 9e-17
Identities = 47/123 (38%), Positives = 70/123 (56%), Gaps = 9/123 (7%)
Frame = +2
Query: 1 MDHAEILQIEPAELTFLTE--KKDFCKLHLINKTTNYVAFRLRLSKPEKYGVTPREGGII 58
M E+LQI P EL F E K+ C L L NK+ NY+AF+++ + P+KY V P G++
Sbjct: 65 MSSGELLQIHPQELQFPFELRKQISCSLQLSNKSDNYIAFKVKTTNPKKYCVRP-NNGVV 241
Query: 59 MPQGTYDIIVSRKSDKLPPPGFECTDRFFIQSMALS----VKDLFTENRGKR---HVEEK 111
+P+ T DI V+ + K PP +C D+F +QS+ S KD+ E K V+E
Sbjct: 242 LPRSTCDITVTMQGQKEAPPDMQCKDKFLLQSVVASPGATAKDITPEMFNKESGYEVDEC 421
Query: 112 ELK 114
+L+
Sbjct: 422 KLR 430
>TC78518 similar to PIR|JC7234|JC7234 27k vesicle-associated membrane
protein-associated protein - curled-leaved tobacco,
partial (83%)
Length = 827
Score = 78.2 bits (191), Expect = 4e-16
Identities = 47/123 (38%), Positives = 71/123 (57%), Gaps = 9/123 (7%)
Frame = +1
Query: 1 MDHAEILQIEPAELTFLTE--KKDFCKLHLINKTTNYVAFRLRLSKPEKYGVTPREGGII 58
M E+L I+P EL F E K+ C L L NKT NYVAF+++ + P+KY V P G++
Sbjct: 82 MSTGELLNIQPDELQFPFELRKQISCSLQLSNKTDNYVAFKVKTTNPKKYCVRP-NSGVV 258
Query: 59 MPQGTYDIIVSRKSDKLPPPGFECTDRFFIQSMALS----VKDLFTENRGK---RHVEEK 111
+P+ T D+ V+ ++ K P +C D+F +QS+ S KD+ E K +VEE
Sbjct: 259 LPRSTCDVTVTMQAQKEAPSDMQCKDKFLLQSVIASPGTTTKDINPEMFSKEAGHNVEEC 438
Query: 112 ELK 114
+L+
Sbjct: 439 KLR 447
>TC81745 similar to PIR|JC7234|JC7234 27k vesicle-associated membrane
protein-associated protein - curled-leaved tobacco,
partial (94%)
Length = 945
Score = 77.4 bits (189), Expect = 8e-16
Identities = 45/115 (39%), Positives = 69/115 (59%), Gaps = 8/115 (6%)
Frame = +2
Query: 5 EILQIEPAELTFLTE--KKDFCKLHLINKTTNYVAFRLRLSKPEKYGVTPREGGIIMPQG 62
++L IEP EL F E K+ C L L NKT +YVAF+++ + P KY V P GI++P+
Sbjct: 116 DLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNT-GIVLPRS 292
Query: 63 TYDIIVSRKSDKLPPPGFECTDRFFIQSM----ALSVKDLFTE--NRGKRHVEEK 111
T D++V+ ++ K P +C D+F +QS+ +S KD+ E N+ HV E+
Sbjct: 293 TCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEE 457
>BI266722 similar to GP|17473763|gb| putative protein {Arabidopsis thaliana},
partial (28%)
Length = 404
Score = 76.3 bits (186), Expect = 2e-15
Identities = 38/100 (38%), Positives = 61/100 (61%), Gaps = 6/100 (6%)
Frame = +2
Query: 1 MDHAEILQIEPAELTFL------TEKKDFCKLHLINKTTNYVAFRLRLSKPEKYGVTPRE 54
M ++L IEP EL FL +K+ C L L NKT +YVAF+++ + P KY V P
Sbjct: 104 MSTGDLLNIEPVELKFLFSVSVELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNT 283
Query: 55 GGIIMPQGTYDIIVSRKSDKLPPPGFECTDRFFIQSMALS 94
G+++PQ T ++IV+ ++ K PP +C D+ +QS+ ++
Sbjct: 284 -GVVLPQSTCEVIVTMQAQKEVPPDMQCKDKXLLQSVRVN 400
>AW775086 similar to PIR|JC7234|JC7 27k vesicle-associated membrane
protein-associated protein - curled-leaved tobacco,
partial (52%)
Length = 747
Score = 68.2 bits (165), Expect = 5e-13
Identities = 36/87 (41%), Positives = 53/87 (60%), Gaps = 2/87 (2%)
Frame = +2
Query: 7 LQIEPAELTFLTEKKDFCKL--HLINKTTNYVAFRLRLSKPEKYGVTPREGGIIMPQGTY 64
LQI P EL F E K + + NK+ NY+AF+++ + PEKY V P G +++P+ T
Sbjct: 44 LQIHPQELHFPFELKKQISISFQMSNKSDNYLAFKVKTTIPEKYCVRPNNG-VVLPRSTC 220
Query: 65 DIIVSRKSDKLPPPGFECTDRFFIQSM 91
DI V+ ++ K PP C D+F IQS+
Sbjct: 221 DIQVTMQAQKKAPPNMHCIDKFLIQSI 301
>BG452644 similar to GP|21554053|gb putative VAMP-associated protein
{Arabidopsis thaliana}, partial (61%)
Length = 629
Score = 67.8 bits (164), Expect = 6e-13
Identities = 46/146 (31%), Positives = 72/146 (48%), Gaps = 35/146 (23%)
Frame = +2
Query: 1 MDHAEILQIEPAELTFLTE--KKDFCKLHLINKTTNYVAFRLRLSKPEKYGVTPREGGII 58
M ++L IEP EL FL E K+ C L L NKT +YVAF+++ + P KY V P G++
Sbjct: 80 MSTGDLLNIEPVELKFLFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNT-GVV 256
Query: 59 MPQGTYDI---------------------------IVSRKSDKLPPPGFECTDRFFIQSM 91
+PQ T ++ IV+ ++ K PP +C D+F +QS+
Sbjct: 257 LPQSTCEVIGIACYIIFSFLF*FFAG*WHCDLWLFIVTMQAQKEVPPDMQCKDKFLLQSV 436
Query: 92 ALS----VKDLFTE--NRGKRHVEEK 111
++ K++ E N+ HV E+
Sbjct: 437 RVNDGADAKEITPEMFNKEAGHVVEE 514
>CA917079 weakly similar to PIR|B86220|B86 protein F22O13.31 [imported] -
Arabidopsis thaliana, partial (23%)
Length = 487
Score = 51.6 bits (122), Expect = 4e-08
Identities = 27/87 (31%), Positives = 50/87 (57%), Gaps = 8/87 (9%)
Frame = +1
Query: 36 VAFRLRLSKPEKYGVTPREGGIIMPQGTYDIIVSRKSDKLPPPGFECTDRFFIQSMALSV 95
VAF+++ + P+KY V P G++ P+ T + IV+ ++ ++ P C D+F +QS +
Sbjct: 1 VAFKVKTTSPKKYSVRPNV-GVLAPKATSEFIVTMQAQRVAPEDLVCKDKFLVQSTLVDA 177
Query: 96 K--------DLFTENRGKRHVEEKELK 114
+ LF ++ G R++EE +LK
Sbjct: 178 EITTEDVTSSLFVKD-GLRYIEENKLK 255
>TC83451 weakly similar to GP|19310653|gb|AAL85057.1 unknown protein
{Arabidopsis thaliana}, partial (33%)
Length = 558
Score = 28.1 bits (61), Expect = 0.53
Identities = 20/71 (28%), Positives = 31/71 (43%)
Frame = +3
Query: 26 LHLINKTTNYVAFRLRLSKPEKYGVTPREGGIIMPQGTYDIIVSRKSDKLPPPGFECTDR 85
L L+ T +V L L+ G+ PR +P YD S S++ PPP +
Sbjct: 249 LILVILLTIFVLIALILTSGRDPGIVPRNSYPPLPDN-YDGSDSNNSEQNPPPHLPRSKE 425
Query: 86 FFIQSMALSVK 96
+ +A+ VK
Sbjct: 426 VIVNGIAVRVK 458
>TC89468 similar to GP|5305145|emb|CAB46084.1 fructose-1 6-bisphosphatase
{Pisum sativum}, partial (94%)
Length = 1161
Score = 26.6 bits (57), Expect = 1.5
Identities = 11/36 (30%), Positives = 20/36 (55%)
Frame = -1
Query: 2 DHAEILQIEPAELTFLTEKKDFCKLHLINKTTNYVA 37
+H + L + P + ++ DFC+ L+N+ TN A
Sbjct: 186 NHIQFLLLLPLNIGVSSKS*DFCQSSLVNRRTNKFA 79
>TC86808 homologue to SP|Q9ZTR1|SPD1_PEA Spermidine synthase 1 (EC 2.5.1.16)
(Putrescine aminopropyltransferase 1) (SPDSY 1). [Garden
pea], partial (93%)
Length = 1350
Score = 26.2 bits (56), Expect = 2.0
Identities = 12/37 (32%), Positives = 23/37 (61%)
Frame = +1
Query: 60 PQGTYDIIVSRKSDKLPPPGFECTDRFFIQSMALSVK 96
P+GTYD ++ SD + P E ++ F +S+A +++
Sbjct: 658 PEGTYDAVIVDSSDPI-GPAQELFEKPFFESVARALR 765
>BI311333
Length = 706
Score = 25.4 bits (54), Expect = 3.4
Identities = 15/52 (28%), Positives = 26/52 (49%)
Frame = +2
Query: 20 KKDFCKLHLINKTTNYVAFRLRLSKPEKYGVTPREGGIIMPQGTYDIIVSRK 71
K+ C L K++ R+ L KPE PR+ +I +G + + VS++
Sbjct: 56 KRYRCDSKLNGKSSMLAHCRVYLKKPENMLNDPRQTNLINGKGDFLVSVSQR 211
>TC84305 weakly similar to PIR|T39881|T39881 preg-like protein - fission
yeast (Schizosaccharomyces pombe), partial (16%)
Length = 741
Score = 25.4 bits (54), Expect = 3.4
Identities = 12/42 (28%), Positives = 18/42 (42%)
Frame = -1
Query: 41 RLSKPEKYGVTPREGGIIMPQGTYDIIVSRKSDKLPPPGFEC 82
R S + PR+G ++ P +Y +DK FEC
Sbjct: 660 RRSSKQHIQFQPRDGSLLSPSNSYRSRTLTPADKTQVAPFEC 535
>TC80730 weakly similar to GP|20198248|gb|AAD26886.2 expressed protein
{Arabidopsis thaliana}, partial (27%)
Length = 827
Score = 25.4 bits (54), Expect = 3.4
Identities = 11/23 (47%), Positives = 15/23 (64%)
Frame = -3
Query: 77 PPGFECTDRFFIQSMALSVKDLF 99
PPGF C+ +FFI S + +LF
Sbjct: 465 PPGFSCSKKFFI-SFFICFSNLF 400
>TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 kDa protein
2. [Tomato] {Lycopersicon esculentum}, complete
Length = 2278
Score = 25.0 bits (53), Expect = 4.4
Identities = 17/51 (33%), Positives = 25/51 (48%)
Frame = -3
Query: 28 LINKTTNYVAFRLRLSKPEKYGVTPREGGIIMPQGTYDIIVSRKSDKLPPP 78
L+N T + L S P K V+P + + IVSR++ K+PPP
Sbjct: 893 LLNS*TKWFTILLSKSSPPKC-VSP---AVALTSKIPSSIVSRETSKVPPP 753
>TC87385 similar to GP|23093788|gb|AAF50220.2 CG16711-PA {Drosophila
melanogaster}, partial (1%)
Length = 879
Score = 24.6 bits (52), Expect = 5.8
Identities = 10/23 (43%), Positives = 13/23 (56%)
Frame = -1
Query: 9 IEPAELTFLTEKKDFCKLHLINK 31
I LT+ T K F LH++NK
Sbjct: 828 ISSLNLTYTTNKNKFNNLHILNK 760
>BI265380 weakly similar to GP|17932898|em isoamylase {Triticum aestivum},
partial (13%)
Length = 604
Score = 24.3 bits (51), Expect = 7.6
Identities = 11/21 (52%), Positives = 15/21 (71%), Gaps = 2/21 (9%)
Frame = +3
Query: 39 RLRLSK--PEKYGVTPREGGI 57
+L+LSK P YG TP+E G+
Sbjct: 264 QLQLSKGHPSPYGATPQEDGV 326
>TC88441 similar to GP|14194139|gb|AAK56264.1 AT3g06960/F17A9_11
{Arabidopsis thaliana}, partial (47%)
Length = 1798
Score = 24.3 bits (51), Expect = 7.6
Identities = 10/21 (47%), Positives = 16/21 (75%)
Frame = -2
Query: 17 LTEKKDFCKLHLINKTTNYVA 37
LTE+ +FCKL+ +K TN ++
Sbjct: 396 LTEETNFCKLN*PSKNTNQLS 334
>TC86659 similar to GP|5360263|dbj|BAA81904.1 NtPRp27 {Nicotiana tabacum},
partial (83%)
Length = 875
Score = 24.3 bits (51), Expect = 7.6
Identities = 9/23 (39%), Positives = 16/23 (69%)
Frame = +1
Query: 83 TDRFFIQSMALSVKDLFTENRGK 105
+D+FF+Q + +V L+TE + K
Sbjct: 646 SDQFFVQLLGKTVNQLWTEYKAK 714
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.139 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,147,556
Number of Sequences: 36976
Number of extensions: 35703
Number of successful extensions: 154
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 148
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 148
length of query: 114
length of database: 9,014,727
effective HSP length: 90
effective length of query: 24
effective length of database: 5,686,887
effective search space: 136485288
effective search space used: 136485288
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 50 (23.9 bits)
Lotus: description of TM0336.10


