

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
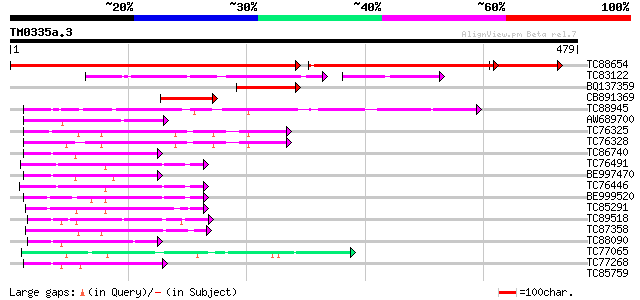
Query= TM0335a.3
(479 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88654 similar to PIR|H84747|H84747 probable steroid dehydrogen... 422 0.0
TC83122 similar to PIR|C84658|C84658 3-beta-hydroxysteroid dehyd... 119 1e-33
BQ137359 weakly similar to PIR|H84747|H847 probable steroid dehy... 81 8e-16
CB891369 78 7e-15
TC88945 similar to PIR|T05987|T05987 hypothetical protein F17M5.... 56 3e-08
AW689700 similar to GP|20196859|gb putative sterol dehydrogenase... 49 4e-06
TC76325 similar to PIR|C96814|C96814 hypothetical protein T30F21... 49 6e-06
TC76328 homologue to PIR|C96814|C96814 hypothetical protein T30F... 49 6e-06
TC86740 weakly similar to PIR|D84747|D84747 probable cinnamoyl-C... 47 2e-05
TC76491 similar to SP|O65781|GAE2_CYATE UDP-glucose 4-epimerase ... 46 3e-05
BE997470 similar to PIR|D84747|D84 probable cinnamoyl-CoA reduct... 46 3e-05
TC76446 similar to SP|O65781|GAE2_CYATE UDP-glucose 4-epimerase ... 46 4e-05
BE999520 homologue to SP|O65781|GAE2 UDP-glucose 4-epimerase GEP... 45 8e-05
TC85291 similar to SP|Q43070|GAE1_PEA UDP-glucose 4-epimerase (E... 44 1e-04
TC89518 weakly similar to GP|23495823|dbj|BAC20033. putative NAD... 43 3e-04
TC87358 similar to GP|13272475|gb|AAK17176.1 unknown protein {Ar... 43 3e-04
TC88090 similar to PIR|F96709|F96709 probable reductase T26J14.1... 43 3e-04
TC77065 similar to PIR|A85356|A85356 nucleotide sugar epimerase-... 42 4e-04
TC77268 weakly similar to GP|10177026|dbj|BAB10264. dihydroflavo... 42 4e-04
TC85759 similar to PIR|T48072|T48072 dTDP-glucose 4-6-dehydratas... 41 0.001
>TC88654 similar to PIR|H84747|H84747 probable steroid dehydrogenase
[imported] - Arabidopsis thaliana, partial (82%)
Length = 1435
Score = 422 bits (1085), Expect(3) = 0.0
Identities = 205/245 (83%), Positives = 220/245 (89%)
Frame = +1
Query: 1 MHLSENEGIEGKTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVR 60
MHLSENEGIEGK+ VVTGGLGFVGSALCLEL+RRGA +VRAFDLR SSPW +LK GV
Sbjct: 31 MHLSENEGIEGKSFVVTGGLGFVGSALCLELIRRGAQQVRAFDLRQSSPWSHLLKLKGVN 210
Query: 61 CIQGDIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLG 120
CIQGD+ RKEDVE LRGADCVFHLAAFGMSGKEMLQ RVD+VNI GTCH+LDAC+ LG
Sbjct: 211 CIQGDVTRKEDVERVLRGADCVFHLAAFGMSGKEMLQFGRVDEVNINGTCHILDACIDLG 390
Query: 121 IKRLVYCSTYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKND 180
IKRLVYCSTYNVVF GQ+I+NGNE+LPYFPID HVDPY RSKSIAEQLVLKNNAR KND
Sbjct: 391 IKRLVYCSTYNVVFGGQKILNGNEALPYFPIDRHVDPYSRSKSIAEQLVLKNNARTLKND 570
Query: 181 AGNCLYTCAVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALI 240
N LYTCAVRPAAIYGPGEDRHLPRIIT A+LGLLLF IGD+TVKSDWVFV+NLVLALI
Sbjct: 571 TRNHLYTCAVRPAAIYGPGEDRHLPRIITMARLGLLLFRIGDKTVKSDWVFVDNLVLALI 750
Query: 241 LASMG 245
+ASMG
Sbjct: 751 MASMG 765
Score = 274 bits (701), Expect(3) = 0.0
Identities = 134/162 (82%), Positives = 147/162 (90%), Gaps = 1/162 (0%)
Frame = +3
Query: 253 KGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELPKTSLAVDHALVLGRICQGVYT 312
KGK RPIAAGQAYFI DGSPVN+FEFLQPLLRSL+Y+LPK SLA++HALVL +ICQ VYT
Sbjct: 789 KGK-RPIAAGQAYFICDGSPVNSFEFLQPLLRSLDYDLPKRSLALEHALVLAKICQRVYT 965
Query: 313 ILYPWLDRWWLPQPFIL-PSEVHKVGVTHYFSYLKAKEEIGYVPMVSSREGMASTISYWQ 371
ILYP L+RWWLPQPFIL PSE KVGVTHYFSYLKAKEE+GYVPMV+SREGM STISYW+
Sbjct: 966 ILYPLLNRWWLPQPFILLPSEALKVGVTHYFSYLKAKEELGYVPMVTSREGMDSTISYWK 1145
Query: 372 QRKMITLDGPTIYTWLFCVIGMISLFCAAFLPDVGIVFLLRA 413
QRK LDGPTIYTWLFCV+GM SLFCA FLPD+GI+FLLRA
Sbjct: 1146QRKRQILDGPTIYTWLFCVVGMTSLFCAGFLPDMGIMFLLRA 1271
Score = 96.7 bits (239), Expect(3) = 0.0
Identities = 44/62 (70%), Positives = 47/62 (74%)
Frame = +1
Query: 406 GIVFLLRATSLFVFRSMWMIRLVFILATAAHVFEAIYAWYLAKRVDHANARGWFWQTFAL 465
G+ F LFVFRSMWM RLVFI+ATA H EAIYAWYL KRVD NARGW WQTFAL
Sbjct: 1249 GLCFFSEL*CLFVFRSMWMTRLVFIIATAVHFIEAIYAWYLTKRVDPVNARGWLWQTFAL 1428
Query: 466 GY 467
G+
Sbjct: 1429 GF 1434
>TC83122 similar to PIR|C84658|C84658 3-beta-hydroxysteroid dehydrogenase
[imported] - Arabidopsis thaliana, partial (76%)
Length = 1102
Score = 119 bits (298), Expect(2) = 1e-33
Identities = 81/205 (39%), Positives = 113/205 (54%), Gaps = 1/205 (0%)
Frame = +2
Query: 65 DIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRL 124
D+ K + A G + VFH+AA S QL VN+ GT +V+DAC+ L +KRL
Sbjct: 17 DLRNKPQLLKAFDGVEVVFHMAAPNSSINNY-QLHH--SVNVEGTKNVIDACVELKVKRL 187
Query: 125 VYCSTYNVVFAG-QRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAGN 183
+Y S+ +VVF G I NG+ESLPY P H D Y +K+ E LV+K N +
Sbjct: 188 IYTSSPSVVFDGIHGIHNGSESLPYPP--SHNDHYSATKAEGEGLVIKANGT-------S 340
Query: 184 CLYTCAVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALILAS 243
L TC +RP++I+GPG+ +P ++ AK G F +GD D+ +VEN+ A I A
Sbjct: 341 GLLTCCIRPSSIFGPGDKLLVPSLVDAAKAGKSKFIVGDGNNVYDFTYVENVAHAHICAD 520
Query: 244 MGLLDDSAGKGKQRPIAAGQAYFIS 268
L A +G AAG+AYFI+
Sbjct: 521 RAL----ASEGTVSEKAAGEAYFIT 583
Score = 42.4 bits (98), Expect(2) = 1e-33
Identities = 22/86 (25%), Positives = 41/86 (47%)
Frame = +1
Query: 282 LLRSLEYELPKTSLAVDHALVLGRICQGVYTILYPWLDRWWLPQPFILPSEVHKVGVTHY 341
+L L Y+ P + V + + + + +Y +L P+ + P + PS + T
Sbjct: 625 ILEGLGYQRPSIKIPVFVIMPIAHLVEWIYRLLGPY----GMKVPQLTPSRIRLTSCTRS 792
Query: 342 FSYLKAKEEIGYVPMVSSREGMASTI 367
F KAK+ + Y P++ +EG+ TI
Sbjct: 793 FDCSKAKDRLDYAPIIPLQEGIRRTI 870
>BQ137359 weakly similar to PIR|H84747|H847 probable steroid dehydrogenase
[imported] - Arabidopsis thaliana, partial (10%)
Length = 956
Score = 81.3 bits (199), Expect = 8e-16
Identities = 40/54 (74%), Positives = 47/54 (86%)
Frame = +2
Query: 192 PAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALILASMG 245
PAAIYGPGE +HLPRIIT A+LGLLL+ +GD+TVKS VFV+NLVLALI A+ G
Sbjct: 74 PAAIYGPGEHKHLPRIITMARLGLLLYKMGDKTVKSHRVFVDNLVLALIKANSG 235
>CB891369
Length = 155
Score = 78.2 bits (191), Expect = 7e-15
Identities = 36/48 (75%), Positives = 42/48 (87%)
Frame = +3
Query: 128 STYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNAR 175
STY VV+ GQ+I+NGNE+LPY PID HV+PY RSKS+AEQ VLKNNAR
Sbjct: 6 STYKVVYGGQKILNGNEALPYPPIDRHVEPYSRSKSMAEQ*VLKNNAR 149
>TC88945 similar to PIR|T05987|T05987 hypothetical protein F17M5.120 -
Arabidopsis thaliana, partial (94%)
Length = 1345
Score = 56.2 bits (134), Expect = 3e-08
Identities = 92/396 (23%), Positives = 160/396 (40%), Gaps = 9/396 (2%)
Frame = +3
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIVRKED 71
K +VVTG G++G LC L R+G + V+ +R +S A+ + + GDI
Sbjct: 105 KKVVVTGASGYLGGKLCNSLHRQG-YSVKVI-VRPTSNLSALPPSTEI--VYGDITDFSS 272
Query: 72 VESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHL-GIKRLVYCSTY 130
+ SA VFHLAA + + ++ VN+ G +VL+A +++LVY S++
Sbjct: 273 LLSAFSDCSVVFHLAA--LVEPWLPDPSKFITVNVEGLKNVLEAVKQTKTVEKLVYTSSF 446
Query: 131 NVVFAGQRIVNGNESLPYFPIDHH----VDPYGRSKSIAEQLVLKNNARPFKNDAGNCLY 186
+ + + HH Y +SK +++ L+ A +
Sbjct: 447 FALGPTDGAIADENQV------HHERFFCTEYEKSKVATDKIALQ--------AASEGVP 584
Query: 187 TCAVRPAAIYGPGE---DRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALILAS 243
+ P IYGPG+ + +++ G L IG K + V+++V I A
Sbjct: 585 IVLLYPGVIYGPGKVTAGNVVAKMLVERFSGRLPGYIGKGNDKFSFSHVDDVVEGHIAAM 764
Query: 244 MGLLDDSAGKGKQRPIAAGQAYFIS-DGSPVNTFEFLQPLLRSLEYELPKTSLAVDHALV 302
KG+ G+ Y ++ + + N + ++ + + L V A
Sbjct: 765 K--------KGQ-----IGERYLLTGENASFNQVFDMAAVITNTSKPMVSIPLCVIEA-- 899
Query: 303 LGRICQGVYTILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKEEIGYVPMVSSREG 362
Y L + R PFI P VH + +S KAK E+ Y P S REG
Sbjct: 900 --------YGWLLVLISRITGKLPFISPPTVHVLRHRWEYSCEKAKMELDYKPR-SLREG 1052
Query: 363 MASTISYWQQRKMITLDGPTIYTWLFCVIGMISLFC 398
+A + + + ++ + ++ + GMI C
Sbjct: 1053LAEVLIWLKNLGLVKY*RNVLLLFIGLLFGMIYNTC 1160
>AW689700 similar to GP|20196859|gb putative sterol dehydrogenase
{Arabidopsis thaliana}, partial (21%)
Length = 549
Score = 48.9 bits (115), Expect = 4e-06
Identities = 38/130 (29%), Positives = 60/130 (45%), Gaps = 7/130 (5%)
Frame = +3
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFD------LRDSSPWFA-ILKDNGVRCIQG 64
KT VV GG GF+G +L L+L++ G VR D L S A L +
Sbjct: 102 KTCVVLGGRGFIGKSLVLQLLKLGNWIVRIADSTHSLNLHHSESLLAEALSSSRASYFHL 281
Query: 65 DIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRL 124
D+ K + L G+ VF+ + + + ++ + G +V+ AC +KRL
Sbjct: 282 DLTDKHRIAKVLEGSSVVFYFDVDSSNNGD--HFCSLYKLIVQGAKNVIIACRESKVKRL 455
Query: 125 VYCSTYNVVF 134
+Y S+ +VVF
Sbjct: 456 IYNSSADVVF 485
>TC76325 similar to PIR|C96814|C96814 hypothetical protein T30F21.10
[imported] - Arabidopsis thaliana, partial (97%)
Length = 2416
Score = 48.5 bits (114), Expect = 6e-06
Identities = 60/244 (24%), Positives = 102/244 (41%), Gaps = 17/244 (6%)
Frame = +1
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKD-------NGVRCIQG 64
K +++TG GF+ S + LVR +H + D + + LK+ + ++G
Sbjct: 40 KNILITGAAGFIASHVANRLVR--SHPEYKIVVLDKLDYCSNLKNLLPSKASPNFKFVKG 213
Query: 65 DIVRKEDVESAL--RGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLG-I 121
DI + V L D + H AA + NI GT +L+AC G I
Sbjct: 214 DIGSADLVNYLLITESIDTIMHFAAQTHVDNSFGNSFEFTKNNIYGTHVLLEACKVTGQI 393
Query: 122 KRLVYCSTYNVVFAGQR--IVNGNESLPYFPIDHHVDPYGRSKSIAEQLVL---KNNARP 176
+R ++ ST V +V +E+ P +PY +K+ AE LV+ ++ P
Sbjct: 394 RRFIHVSTDEVYGETDEDAVVGNHEASQLLP----TNPYSATKAGAEMLVMAYGRSYGLP 561
Query: 177 FKNDAGNCLYTCAVRPAAIYGPGE--DRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVEN 234
GN +YGP + ++ +P+ I A G L GD + +++ E+
Sbjct: 562 VITTRGN----------NVYGPNQFPEKLIPKFILLAMQGKTLPIHGDGSNVRSYLYCED 711
Query: 235 LVLA 238
+ A
Sbjct: 712 VAEA 723
>TC76328 homologue to PIR|C96814|C96814 hypothetical protein T30F21.10
[imported] - Arabidopsis thaliana, partial (56%)
Length = 1492
Score = 48.5 bits (114), Expect = 6e-06
Identities = 63/250 (25%), Positives = 102/250 (40%), Gaps = 23/250 (9%)
Frame = +2
Query: 12 KTLVVTGGLGFVGSALCLELVRR-GAHEVRAFDLRD------------SSPWFAILKDNG 58
K +++TG GF+ S + L+R +++ D D SSP F +K
Sbjct: 128 KNILITGAAGFIASHVANRLIRNYPDYKIVVLDKLDYCSNLKNLIPSRSSPNFKFVK--- 298
Query: 59 VRCIQGDIVRKEDVESAL--RGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDAC 116
GDI + V L D + H AA + NI GT +L+AC
Sbjct: 299 -----GDIESADLVNYLLITESIDTIMHFAAQTHVDNSFGNSFEFTKNNIYGTHVLLEAC 463
Query: 117 LHLG-IKRLVYCSTYNVVFAGQR--IVNGNESLPYFPIDHHVDPYGRSKSIAEQLVL--- 170
G IKR ++ ST V +V +E+ P +PY +K+ AE LV+
Sbjct: 464 KVTGQIKRFIHVSTDEVYGETDEDAVVGNHEASQLLP----TNPYSATKAGAEMLVMAYG 631
Query: 171 KNNARPFKNDAGNCLYTCAVRPAAIYGPGE--DRHLPRIITTAKLGLLLFTIGDQTVKSD 228
++ P GN +YGP + ++ +P+ I A G +L GD +
Sbjct: 632 RSYGLPVITTRGN----------NVYGPNQFPEKLIPKFILLAMQGKVLPIHGDGSNVRS 781
Query: 229 WVFVENLVLA 238
+++ E++ A
Sbjct: 782 YLYCEDVAEA 811
>TC86740 weakly similar to PIR|D84747|D84747 probable cinnamoyl-CoA
reductase [imported] - Arabidopsis thaliana, partial
(85%)
Length = 1198
Score = 46.6 bits (109), Expect = 2e-05
Identities = 38/123 (30%), Positives = 58/123 (46%), Gaps = 5/123 (4%)
Frame = +1
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAIL-----KDNGVRCIQGDI 66
K + VTG GFV S L L+ +G + V SP + L + + DI
Sbjct: 85 KKVCVTGAGGFVASWLVKLLLSKG-YFVHGTVREPGSPKYEHLLKLEKASENLTLFKADI 261
Query: 67 VRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLVY 126
+ E V SA+ G VFH+A+ S V + + GT +VL+ACL ++R+V+
Sbjct: 262 LDYESVYSAIVGCSAVFHVASPVPSTVVPNPEVEVIEPAVKGTANVLEACLKANVERVVF 441
Query: 127 CST 129
S+
Sbjct: 442 VSS 450
>TC76491 similar to SP|O65781|GAE2_CYATE UDP-glucose 4-epimerase GEPI48 (EC
5.1.3.2) (Galactowaldenase) (UDP- galactose
4-epimerase)., partial (96%)
Length = 1805
Score = 46.2 bits (108), Expect = 3e-05
Identities = 44/169 (26%), Positives = 73/169 (43%), Gaps = 10/169 (5%)
Frame = +1
Query: 10 EGKTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIVRK 69
+ KT++VTGG G++GS L+L+ G V D D+S A+ + + G +
Sbjct: 265 QNKTVLVTGGAGYIGSHTVLQLLLGGFKTV-IVDNLDNSSEVAVRRVKELAAEFGKNLNF 441
Query: 70 EDVESALRGA----------DCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHL 119
V+ + A D V H A G+ + + N+ GT +L+
Sbjct: 442 HKVDLRDKAALEQIFSSTKFDAVIHFAGMKAVGESVQKPLLYYNNNLIGTITLLEVMAAH 621
Query: 120 GIKRLVYCSTYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQL 168
G K+LV+ S+ V+ + V E P +PYGR+K E++
Sbjct: 622 GCKKLVFSSS-ATVYGWPKEVPCTEEFPL----SAANPYGRTKLTIEEI 753
>BE997470 similar to PIR|D84747|D84 probable cinnamoyl-CoA reductase
[imported] - Arabidopsis thaliana, partial (44%)
Length = 807
Score = 46.2 bits (108), Expect = 3e-05
Identities = 38/127 (29%), Positives = 60/127 (46%), Gaps = 9/127 (7%)
Frame = +3
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAIL-----KDNGVRCIQGDI 66
K + VTG GFV S L L+ +G + V SP + L + + DI
Sbjct: 87 KKVCVTGAGGFVASWLVKLLLSKG-YFVHGTVREPGSPKYEHLLKLEKASENLTLFKADI 263
Query: 67 VRKEDVESALRGADCVFHLA----AFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIK 122
+ E V SA+ G VFH+A + + E+ V + + GT +VL+ACL ++
Sbjct: 264 LDYESVYSAIVGCSAVFHVASPVPSTVVPNPELNLQVEVIEPAVKGTANVLEACLKANVE 443
Query: 123 RLVYCST 129
R+V+ S+
Sbjct: 444 RVVFVSS 464
>TC76446 similar to SP|O65781|GAE2_CYATE UDP-glucose 4-epimerase GEPI48 (EC
5.1.3.2) (Galactowaldenase) (UDP- galactose
4-epimerase)., partial (55%)
Length = 703
Score = 45.8 bits (107), Expect = 4e-05
Identities = 45/170 (26%), Positives = 73/170 (42%), Gaps = 10/170 (5%)
Frame = +1
Query: 9 IEGKTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIVR 68
+ KT++VTGG G++GS L+L+ G + D D+S AI + + G+ +
Sbjct: 43 MNNKTVLVTGGAGYIGSHTVLQLLLGGFKSI-VVDNLDNSSEVAIHRVKELAGEFGNNLS 219
Query: 69 KEDVESALRGA----------DCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLH 118
V+ R A D V H A G+ + N+ GT +L+
Sbjct: 220 FHKVDLRDRAALEQIFGSTTFDAVIHFAGLKAVGESAQKPLLYYNNNLIGTITLLEVMAA 399
Query: 119 LGIKRLVYCSTYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQL 168
G K+LV+ S+ V+ + V E P +PYGR+K E++
Sbjct: 400 HGCKKLVFSSS-ATVYGWPKEVPCTEEFPL----SAANPYGRTKLTIEEI 534
>BE999520 homologue to SP|O65781|GAE2 UDP-glucose 4-epimerase GEPI48 (EC
5.1.3.2) (Galactowaldenase) (UDP- galactose
4-epimerase)., partial (46%)
Length = 499
Score = 44.7 bits (104), Expect = 8e-05
Identities = 44/170 (25%), Positives = 74/170 (42%), Gaps = 13/170 (7%)
Frame = +3
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIV---- 67
KT++VTGG G++GS L+L+ G ++V D D+S +I + V+ + GD
Sbjct: 3 KTILVTGGAGYIGSHTVLQLL-LGGYKVVVVDNLDNSSQKSI---DRVKKLAGDFAGNLS 170
Query: 68 -------RKEDVESALRGA--DCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLH 118
++ +E D V H A G+ + + N+ GT + +
Sbjct: 171 FHKLDLRDRDGLEKIFSSTKFDAVIHFAGLKAVGESVQKPLLYYDNNLIGTIVLFEVMAA 350
Query: 119 LGIKRLVYCSTYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQL 168
G K+LV+ S+ V+ + V E P PYGR+K E++
Sbjct: 351 HGCKKLVFSSS-ATVYGWPKEVPCTEEFPL----SAASPYGRTKLYIEEI 485
>TC85291 similar to SP|Q43070|GAE1_PEA UDP-glucose 4-epimerase (EC 5.1.3.2)
(Galactowaldenase) (UDP- galactose 4-epimerase). [Garden
pea], partial (78%)
Length = 1018
Score = 44.3 bits (103), Expect = 1e-04
Identities = 41/166 (24%), Positives = 72/166 (42%), Gaps = 11/166 (6%)
Frame = +1
Query: 14 LVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILK---------DNGVRCIQG 64
++VTGG GF+G+ L+L+ +G V D D+S A+ + + G
Sbjct: 208 ILVTGGSGFIGTHTVLQLL-QGGFAVSIIDNFDNSVIAAVDRVRELVGPQLSQNLDFTLG 384
Query: 65 DIVRKEDVESALRGA--DCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIK 122
D+ K+D+E D V H A G+ + R N+ GT ++ + K
Sbjct: 385 DLRIKDDLEKLFSKTKFDAVIHFAGLKAVGESVANPRRYFDNNLVGTINLYEVMAKYNCK 564
Query: 123 RLVYCSTYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQL 168
++V+ S+ V I + F + ++PYGR+K E++
Sbjct: 565 KMVFSSSATVYGQPDTI----PCVEDFKL-QAMNPYGRTKLFLEEI 687
>TC89518 weakly similar to GP|23495823|dbj|BAC20033. putative NADPH HC toxin
reductase {Oryza sativa (japonica cultivar-group)},
partial (28%)
Length = 828
Score = 42.7 bits (99), Expect = 3e-04
Identities = 46/179 (25%), Positives = 87/179 (47%), Gaps = 22/179 (12%)
Frame = +3
Query: 16 VTGGLGFVGSALCLELVRRGAHEVRAF--DLRDSSPWFAILK-----DNGVRCIQGDIVR 68
VTGG G++GS L +L+ +G + V A +L+D S + L+ + + + DI +
Sbjct: 51 VTGGAGYIGSLLVKKLLEKG-YTVHATLRNLKDESK-VSFLRGFPHANTRLVLFEADIYK 224
Query: 69 KEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACL-HLGIKRLVYC 127
+D +A++G + VFH+A + + Q +++ I G + C+ ++RL+Y
Sbjct: 225 PDDFGTAIQGCEFVFHVATPYLHQTDS-QFRSIEEAAIAGVKSIAATCIKSRTVRRLIYT 401
Query: 128 STYNVVFAGQRIVNGN--------------ESLPYFPIDHHVDPYGRSKSIAEQLVLKN 172
T VV A +G+ +SL + P+ Y SK++AE+ +L++
Sbjct: 402 GT--VVAASPLKDDGSGYKDFIDETCWTPLQSL-HLPLTDFHKGYVASKTLAERELLRS 569
>TC87358 similar to GP|13272475|gb|AAK17176.1 unknown protein {Arabidopsis
thaliana}, partial (92%)
Length = 1688
Score = 42.7 bits (99), Expect = 3e-04
Identities = 41/165 (24%), Positives = 71/165 (42%), Gaps = 8/165 (4%)
Frame = +2
Query: 14 LVVTGGLGFVGSALCLELVRRGAHEVRAFDL-RDSSPWFAILKD-----NGVRCIQGDIV 67
++VTGG G++GS L++ +L R + +L+D ++ I D+
Sbjct: 476 VLVTGGAGYIGSHAAFRLLKDNYRVTIVDNLSRGNLGAVRVLQDLFPEPGRLQFIYADLG 655
Query: 68 RKEDVESAL--RGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLV 125
K+ V D V H AA G+ + + + T VL++ +K L+
Sbjct: 656 DKKSVNKIFLENKFDAVMHFAAVAYVGESTVDPLKYYHNITSNTLLVLESMAKHDVKTLI 835
Query: 126 YCSTYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVL 170
Y ST + + E P PI +PYG++K +AE ++L
Sbjct: 836 YSSTC-ATYGEPEKMPITEVTPQVPI----NPYGKAKKMAEDIIL 955
>TC88090 similar to PIR|F96709|F96709 probable reductase T26J14.11
[imported] - Arabidopsis thaliana, complete
Length = 1284
Score = 42.7 bits (99), Expect = 3e-04
Identities = 36/121 (29%), Positives = 61/121 (49%), Gaps = 7/121 (5%)
Frame = +1
Query: 16 VTGGLGFVGSALCLELVRRGAHEVRAF-----DLRDSSPWFAILKD-NGVRCIQGDIVRK 69
VTGG GF+ + L L+ +G H VR DL + D ++ ++ D++ +
Sbjct: 100 VTGGTGFIAAYLVKALLEKG-HIVRTTVRNPDDLEKVGYLTQLSGDKERLKILKADLMVE 276
Query: 70 EDVESALRGADCVFHLAA-FGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLVYCS 128
+ A+ G D VFH A+ + +Q T +D I GT +VL++C+ +KR+V S
Sbjct: 277 GSFDEAVTGVDGVFHTASPVIVPYDNNIQATLIDPC-IKGTQNVLNSCIKANVKRVVLTS 453
Query: 129 T 129
+
Sbjct: 454 S 456
>TC77065 similar to PIR|A85356|A85356 nucleotide sugar epimerase-like
protein [imported] - Arabidopsis thaliana, partial (97%)
Length = 1679
Score = 42.4 bits (98), Expect = 4e-04
Identities = 75/303 (24%), Positives = 119/303 (38%), Gaps = 21/303 (6%)
Frame = +2
Query: 11 GKTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRD------SSPWFAILKDNGVRCIQG 64
G +++VTG GF+GS + L L RRG V + D A+L+ GV + G
Sbjct: 362 GMSVLVTGAAGFIGSHVSLALKRRGDGVVGLDNFNDYYDPSLKKARKALLQSRGVFIVHG 541
Query: 65 DIVRKEDVESALRGADC-----VFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHL 119
D+ D + + D V HLAA M NI G +L+AC
Sbjct: 542 DL---NDAKLLAKLFDVVAFTHVMHLAAQAGVRYAMENPMSYVNSNIAGLVTLLEACKTA 712
Query: 120 GIKRLVYCSTYNVVFAGQRIVNG-NESLPYFPIDHHVDP---YGRSKSIAEQLVLKNNAR 175
+ ++V+A V G NE +P+ D P Y +K E++
Sbjct: 713 NPQP-------SIVWASSSSVYGLNEKVPFSESDRTDQPASLYAATKKAGEEI-----TH 856
Query: 176 PFKNDAGNCLYTCAVRPAAIYGP-GEDRHLPRIITTAKLGLLLFTI--GDQTV--KSDWV 230
+ + G L +R +YGP G T L T+ G V D+
Sbjct: 857 TYNHIYG--LSITGLRFFTVYGPWGRPDMAIFTFTRNMLQGKPITVYRGKNRVDLSRDFT 1030
Query: 231 FVENLVLALILASMGLLDDSAGK-GKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYE 289
+++++V + S+ S G GK+R A + + + + SPV + L R L+ +
Sbjct: 1031YIDDIVKGCV-GSLDTSGKSTGSGGKKRGAAPYRIFNLGNTSPVTVPTLVSILERLLKVK 1207
Query: 290 LPK 292
K
Sbjct: 1208AKK 1216
>TC77268 weakly similar to GP|10177026|dbj|BAB10264. dihydroflavonol
4-reductase-like {Arabidopsis thaliana}, partial (93%)
Length = 1240
Score = 42.4 bits (98), Expect = 4e-04
Identities = 34/128 (26%), Positives = 63/128 (48%), Gaps = 6/128 (4%)
Frame = +2
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAF--DLRDSSPWFAILKDNG----VRCIQGD 65
KT+ VTG G +GS + L+ RG + V A DL D + + G ++ + D
Sbjct: 38 KTVCVTGASGAIGSWVVRLLLERG-YTVHATIQDLEDENETKHLEAMEGAKTRLKFFEMD 214
Query: 66 IVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLV 125
++ + + +A++G V HLA + G+ ++ + I GT +VL G++R+V
Sbjct: 215 LLNSDSIAAAVKGCAGVIHLACPNIIGEVKDPEKQILEPAIQGTVNVLKVAKEAGVERVV 394
Query: 126 YCSTYNVV 133
S+ + +
Sbjct: 395 ATSSISAI 418
>TC85759 similar to PIR|T48072|T48072 dTDP-glucose 4-6-dehydratase homolog
D18 - Arabidopsis thaliana, partial (84%)
Length = 1796
Score = 40.8 bits (94), Expect = 0.001
Identities = 32/119 (26%), Positives = 54/119 (44%), Gaps = 3/119 (2%)
Frame = +3
Query: 10 EGKTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCI---QGDI 66
+G +VVTGG GFVGS L L+ RG + +F K+N + + ++
Sbjct: 510 KGLRIVVTGGAGFVGSHLVDRLMARGDSVIVV------DNFFTGRKENVMHHFGNPRFEL 671
Query: 67 VRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLV 125
+R + VE L D ++HLA + + N+ GT ++L +G + L+
Sbjct: 672 IRHDVVEPLLLEVDQIYHLACPASPVHYKFNPVKTIKTNVVGTLNMLGLAKRVGARFLL 848
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.142 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,043,830
Number of Sequences: 36976
Number of extensions: 241399
Number of successful extensions: 1615
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 1554
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1609
length of query: 479
length of database: 9,014,727
effective HSP length: 100
effective length of query: 379
effective length of database: 5,317,127
effective search space: 2015191133
effective search space used: 2015191133
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0335a.3


