

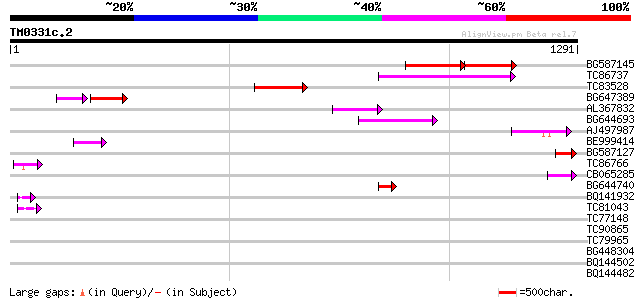
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0331c.2
(1291 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 171 3e-70
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 147 2e-35
TC83528 96 9e-20
BG647389 69 1e-19
AL367832 weakly similar to GP|14091845|gb Putative retroelement ... 86 9e-17
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 83 8e-16
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 69 9e-12
BE999414 59 1e-08
BG587127 weakly similar to PIR|H84506|H84 probable retroelement ... 52 2e-06
TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E... 49 1e-05
CB065285 weakly similar to PIR|A84500|A84 probable retroelement ... 49 2e-05
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 48 2e-05
BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 45 2e-04
TC81043 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproli... 44 5e-04
TC77148 ENOD20 42 0.001
TC90865 homologue to GP|9049326|dbj|BAA99336.1 orf129b {Beta vul... 42 0.002
TC79965 similar to GP|7549635|gb|AAF63820.1| hypothetical protei... 42 0.002
BG448304 41 0.003
BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 41 0.003
BQ144482 similar to GP|6523547|emb| hydroxyproline-rich glycopro... 40 0.004
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 171 bits (433), Expect(2) = 3e-70
Identities = 78/136 (57%), Positives = 105/136 (76%)
Frame = +2
Query: 901 MHPADEDKTAFMTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVK 960
MHP D +KTAF+T R YCY+ MPFGLKNAG+TYQRL++R+FA ++G MEVY+DDM+VK
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 961 SVRGLDHHQDLEEAFGEIRKHNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQ 1020
S+R DH L+E F + ++ M+LNP KC FGV G+FLG+++T +GIE+NP++ AI
Sbjct: 182 SLRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAIL 361
Query: 1021 QMKSPSNVKEVQRLTG 1036
+ SP N +EVQRLTG
Sbjct: 362 DLPSPKNSREVQRLTG 409
Score = 114 bits (285), Expect(2) = 3e-70
Identities = 55/118 (46%), Positives = 80/118 (67%)
Frame = +3
Query: 1036 GRIAALSRFLPKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLKELLSSPPILSKPIQGH 1095
GRIAAL+RF+ +S D+ PF+K L N F W +CEEAF +LK+ L++PP+LSKP G
Sbjct: 408 GRIAALNRFISRSTDKCLPFYKLLCGNKRFVWDEKCEEAFEQLKQYLTTPPVLSKPEAGD 587
Query: 1096 PLHLYFAVSDGALSSVMLQEIDGEHRIVYFVSHTLQGAEVRYQKIEKAALAVLVTARR 1153
L LY A+S A+SSV+++E GE + +++ S + E RY +EK A AV+ +AR+
Sbjct: 588 TLSLYIAISSTAVSSVLIREDRGEQKPIFYTSKRMTDPETRYPTLEKMAFAVITSARK 761
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 147 bits (372), Expect = 2e-35
Identities = 98/317 (30%), Positives = 150/317 (46%), Gaps = 3/317 (0%)
Frame = +1
Query: 839 ANVVIVKKANGKWRMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQ 898
A V+ V+K G R CVDY LN KD YPLP I + +G + +D + H+
Sbjct: 577 APVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETLRRVAGARWFTKLDVVAAFHK 756
Query: 899 IRMHPADEDKTAFMTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMI 958
+R+ D++KTAF T + + PFGL A AT+QR +++ + + Y+DD++
Sbjct: 757 MRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYINKTLHEFLDDFVTAYIDDVL 936
Query: 959 VKSVRG-LDHHQDLEEAFGEIRKHNMRLNPEKCYFGVQGGKFLGFMITS-RGIEINPEKC 1016
+ + DH + + + L+P+KC F V K++GF++T+ +G+ +P K
Sbjct: 937 IYTTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVTTVKYVGFILTAGKGVSCDPLKL 1116
Query: 1017 KAIQQMKSPSNVKEVQRLTGRIAALSRFLPKSGDRSFPFFKCLRKNVAFEWTAECEEAFV 1076
AI+ P +VK + G F+P + + P + RK+ F W AE E AF
Sbjct: 1117 AAIRDWLPPGSVKGARSFLGFCNYYKDFIPGYSEITEPLTRLTRKDFPFRWGAEQEAAFT 1296
Query: 1077 RLKELLSSPPILSKPIQGHPLHLYFAVSDGALSSVMLQEI-DGEHRIVYFVSHTLQGAEV 1135
+LK L + P+L + S AL V+ QE G V F S L AE
Sbjct: 1297 KLKRLFAEEPVLRMFDPEAVTTVETDCSGFALGGVLTQEDGTGAAHPVAFHSQRLSPAEY 1476
Query: 1136 RYQKIEKAALAVLVTAR 1152
Y +K LAV R
Sbjct: 1477 NYPIHDKELLAVWACLR 1527
>TC83528
Length = 555
Score = 95.9 bits (237), Expect = 9e-20
Identities = 53/125 (42%), Positives = 81/125 (64%), Gaps = 4/125 (3%)
Frame = +3
Query: 557 PIVVQLRMNS-FNVRRVLLDQGSSADIIYGDAFDKLGLTDNDLTPYAGTLVGFAGEQVMV 615
P+V++L++N+ F+V RV ++ S DI+Y AF K+ L ++ L P G L G G+ + V
Sbjct: 180 PMVIKLQINNNFSVLRVFVNPMSKVDILYWSAFLKMKLQESMLKPCQGFLKGTFGKGLPV 359
Query: 616 RGYIDLDTIFGEDECARVLKVRYLVLQ---VVASYNVIIGRNTLNRLCAVISTAHLAVKY 672
+GYIDLDT FG+ E + +KVRY V++ V+ YNV++G +L L AV+S A +KY
Sbjct: 360 KGYIDLDTTFGKGENTKTIKVRYFVVESPPSVSIYNVVLGWPSLKDLKAVLSVAEFTIKY 539
Query: 673 PLSSG 677
P+ G
Sbjct: 540 PVGDG 554
>BG647389
Length = 718
Score = 68.6 bits (166), Expect(2) = 1e-19
Identities = 33/85 (38%), Positives = 54/85 (62%)
Frame = +1
Query: 184 KSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYMAR 243
K W+ LPR SI+ +++ + K + QFSA+K+ V+ +L+N+RQ ESL EY+
Sbjct: 451 KEATQQWYMNLPRFSITGYQNMTHKLVHQFSASKHCKVSTTNLFNVRQDPNESLPEYLV* 630
Query: 244 YSAASVKVEDEEPRACALAFKNGLL 268
++ A++KV + + A AF+NGLL
Sbjct: 631 FNNATIKVVNPKQELFAGAFQNGLL 705
Score = 47.8 bits (112), Expect(2) = 1e-19
Identities = 22/70 (31%), Positives = 42/70 (59%)
Frame = +2
Query: 107 RREQRDDGSREADSVAEFRPFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMV 166
++ ++D+ RE + +P S D+ + + +N+K+L L S+ SDP +H FNT+M
Sbjct: 224 QQHEQDELHREDVDELDPQPLSMDIWNAPVLENLKSLSLLSFDDKSDPVEHATTFNTQMA 403
Query: 167 IIAASDAVKC 176
+I A +++C
Sbjct: 404 VIGALKSLRC 433
>AL367832 weakly similar to GP|14091845|gb Putative retroelement {Oryza
sativa}, partial (2%)
Length = 384
Score = 85.9 bits (211), Expect = 9e-17
Identities = 46/118 (38%), Positives = 67/118 (55%), Gaps = 4/118 (3%)
Frame = -1
Query: 736 EETKALKFGD----RTLKIGTRLTEEQETRLTKLLGENLDLFAWSCKDMPGIDPNFICHR 791
EE + + G R +K+G L E + ++ +LL E LD+FA S +DMPG+DP + HR
Sbjct: 354 EEIELINLGTEENKREIKVGAALEEGVKRKIFQLLREYLDIFACSYEDMPGLDPKIVEHR 175
Query: 792 LALNPSLKPVS*LRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVIVKKANG 849
+ P PV * RR D ++ EV K + A F+ V+YP W+AN+V V K +G
Sbjct: 174 IPTKPECPPVR*KLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDG 1
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 82.8 bits (203), Expect = 8e-16
Identities = 58/181 (32%), Positives = 87/181 (48%)
Frame = +2
Query: 794 LNPSLKPVS*LRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVIVKKANGKWRM 853
L P++ P+ R+ K K ++ ++ LL FI+ YP + V+ +KK +G RM
Sbjct: 53 LLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVV-VLFLKKKDGFLRM 229
Query: 854 CVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQIRMHPADEDKTAFMT 913
+DY LN K YPLP ID L D G++ +D G HQ R+ D KTAF
Sbjct: 230 SIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVIGEDVPKTAFRI 409
Query: 914 ARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDLEE 973
+Y M FG N + LM+RVF + + V+ +D+++ S +H L
Sbjct: 410 RYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSKNENEHENHLRL 589
Query: 974 A 974
A
Sbjct: 590 A 592
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 69.3 bits (168), Expect = 9e-12
Identities = 47/167 (28%), Positives = 75/167 (44%), Gaps = 29/167 (17%)
Frame = -2
Query: 1142 KAALAVLVTARRLRPYFQSFPVKVRTDL-PLRQVLQKPDLSGRLVAWSVELSEYSLQYDK 1200
K A+ A+RLR Y + + + + P++ + +KP L+GR+ W + LSEY ++Y
Sbjct: 635 KTCCALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYDIEYRS 456
Query: 1201 RGAVGAQSLADFVV-----ELTPDRFERVDTQ-----------------------WTLFV 1232
+ A+ LAD + + P +F+ D + W L
Sbjct: 455 QKAIKGSILADHLAHQPLEDYRPIKFDFPDEEIMYLKMKDCDEPLFGEGPDPDSVWGLIF 276
Query: 1233 DGSSNSSGSGAGVTLEGPGELVLEQSLKFEFKATNNQAEYEALIAGL 1279
DG+ N G+G G L P + + + F TNN AEYEA I G+
Sbjct: 275 DGAVNVYGNGIGAVLLTPKGAHIPFTARLRFDCTNNIAEYEACIMGI 135
>BE999414
Length = 613
Score = 58.9 bits (141), Expect = 1e-08
Identities = 26/76 (34%), Positives = 41/76 (53%)
Frame = +3
Query: 145 LDSYSGDSDPKDHLLYFNTKMVIIAASDAVKCRMFPSTFKSTAMAWFTTLPRGSISNFRD 204
+DSY G DP +H+ + + AVKC++F +T + AM WF L R SI ++ D
Sbjct: 9 MDSYDGT*DPDEHMENIEVVLTYRSVRGAVKCKLFVTTLRRGAMTWFKNLRRNSIGSWGD 188
Query: 205 FSSKFLVQFSANKNQP 220
+F F+ ++ QP
Sbjct: 189 LCHEFTTHFTVSRTQP 236
>BG587127 weakly similar to PIR|H84506|H84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(13%)
Length = 415
Score = 51.6 bits (122), Expect = 2e-06
Identities = 23/47 (48%), Positives = 34/47 (71%)
Frame = +3
Query: 1244 GVTLEGPGELVLEQSLKFEFKATNNQAEYEALIAGLKLAREVKIRSL 1290
G+ L P +L+QS + EF A+NN+ YEALIAG++LA +KIR++
Sbjct: 3 GIRLTSPTNEILKQSFRLEFHASNNETNYEALIAGVRLAHGLKIRNI 143
>TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E8.80 -
Arabidopsis thaliana, partial (18%)
Length = 1260
Score = 49.3 bits (116), Expect = 1e-05
Identities = 32/77 (41%), Positives = 34/77 (43%), Gaps = 11/77 (14%)
Frame = +2
Query: 10 HSPMRQRISPPRQPHRVVLD-----SPVRTTGVQSPSPP---PSPPVPPP---PPSPSQV 58
HSP SPP PH SP V SP PP P PP PPP PP P
Sbjct: 140 HSPPPPPXSPPPPPHSPTPPVYPYLSPPPPPPVHSPPPPVYSPPPPSPPPCVEPPPPPPP 319
Query: 59 GSLERSPENSPAPEQQP 75
+E P +SPAP Q P
Sbjct: 320 PCVEPPPPSSPAPHQTP 370
Score = 39.7 bits (91), Expect = 0.008
Identities = 24/66 (36%), Positives = 26/66 (39%)
Frame = +2
Query: 10 HSPMRQRISPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSP 69
HSP SPP P V+ P PPP P V PPPPS P SP
Sbjct: 239 HSPPPPVYSPP---------PPSPPPCVEPPPPPPPPCVEPPPPSSPAPHQTPYHPPPSP 391
Query: 70 APEQQP 75
+P P
Sbjct: 392 SPPPSP 409
Score = 35.8 bits (81), Expect = 0.11
Identities = 15/32 (46%), Positives = 17/32 (52%)
Frame = +2
Query: 40 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAP 71
P PPP+P PPPP P S P +SP P
Sbjct: 59 PPPPPAPVFSPPPPVPYYYSSPPPPPAHSPPP 154
Score = 35.4 bits (80), Expect = 0.14
Identities = 21/57 (36%), Positives = 23/57 (39%)
Frame = +2
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 75
PP P V P SP PPP+ PPPP SP P +SP P P
Sbjct: 59 PPPPPAPVFSPPPPVPYYYSSPPPPPAHSPPPPPXSPP-------PPPHSPTPPVYP 208
Score = 33.5 bits (75), Expect = 0.55
Identities = 11/16 (68%), Positives = 13/16 (80%)
Frame = +2
Query: 40 PSPPPSPPVPPPPPSP 55
P PPP+ P PPPPP+P
Sbjct: 32 PPPPPNSPPPPPPPAP 79
Score = 33.1 bits (74), Expect = 0.72
Identities = 21/44 (47%), Positives = 23/44 (51%), Gaps = 11/44 (25%)
Frame = +2
Query: 39 SPSPPPSP----PVPPPP----PSPSQVGSLERSPE---NSPAP 71
SPSPPPSP P PPPP P PS V + P +SP P
Sbjct: 386 SPSPPPSPVYAYPSPPPPVYTSPPPSPVYAYPSPPPPVYSSPPP 517
Score = 32.7 bits (73), Expect = 0.94
Identities = 15/24 (62%), Positives = 17/24 (70%)
Frame = +2
Query: 37 VQSPSPPPSPPVPPPPPSPSQVGS 60
V+SP PPP P PPPPP P+ V S
Sbjct: 20 VRSPPPPP-PNSPPPPPPPAPVFS 88
Score = 30.8 bits (68), Expect = 3.6
Identities = 18/51 (35%), Positives = 19/51 (36%), Gaps = 12/51 (23%)
Frame = +2
Query: 39 SPSPPPSPPVPPP------------PPSPSQVGSLERSPENSPAPEQQPAV 77
SP PPP P PPP PP P V S + P P P V
Sbjct: 143 SPPPPPXSPPPPPHSPTPPVYPYLSPPPPPPVHSPPPPVYSPPPPSPPPCV 295
>CB065285 weakly similar to PIR|A84500|A84 probable retroelement gag/pol
polyprotein [imported] - Arabidopsis thaliana, partial
(2%)
Length = 592
Score = 48.5 bits (114), Expect = 2e-05
Identities = 23/66 (34%), Positives = 39/66 (58%)
Frame = -2
Query: 1225 DTQWTLFVDGSSNSSGSGAGVTLEGPGELVLEQSLKFEFKATNNQAEYEALIAGLKLARE 1284
+++W L DG+ N+ G G G + P + + + F+ TNN AEYEA I G++ A +
Sbjct: 543 NSKWGLVFDGAVNAYGKGIGAVIVSPQGHYIPFTARILFECTNNMAEYEACIFGIEEAID 364
Query: 1285 VKIRSL 1290
++I+ L
Sbjct: 363 MRIKHL 346
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 48.1 bits (113), Expect = 2e-05
Identities = 20/41 (48%), Positives = 28/41 (67%)
Frame = -1
Query: 839 ANVVIVKKANGKWRMCVDYTDLNKACPKDSYPLPSIDSLVD 879
A ++ V+K +G +RMC+DY NK K+ YPLP ID+L D
Sbjct: 238 AALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFD 116
>BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (30%)
Length = 1338
Score = 45.1 bits (105), Expect = 2e-04
Identities = 21/41 (51%), Positives = 24/41 (58%)
Frame = -1
Query: 17 ISPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQ 57
+SPP +P L P SPSPPP PP+PPPPP P Q
Sbjct: 258 VSPPPRPF---LFPPPPNQPSPSPSPPPPPPLPPPPPQPPQ 145
Score = 40.8 bits (94), Expect = 0.003
Identities = 24/67 (35%), Positives = 30/67 (43%), Gaps = 2/67 (2%)
Frame = -1
Query: 12 PMRQRISPPRQPHRVVLDSPVRTT--GVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSP 69
P ++PP PHR T G+ P+ PP PP PPP SP + S P P
Sbjct: 399 PRSSTLAPP--PHRQFHSPSALCTHCGIPPPAVPPPPPPTPPPSSPPALVSPPPRPFLFP 226
Query: 70 APEQQPA 76
P QP+
Sbjct: 225 PPPNQPS 205
Score = 35.4 bits (80), Expect = 0.14
Identities = 28/83 (33%), Positives = 32/83 (37%), Gaps = 8/83 (9%)
Frame = -1
Query: 7 RQHHSPMRQ----RISPPRQPHRVVLDSPVRTTGVQSP----SPPPSPPVPPPPPSPSQV 58
RQ HSP I PP P P T SP SPPP P + PPPP+
Sbjct: 366 RQFHSPSALCTHCGIPPPAVP-----PPPPPTPPPSSPPALVSPPPRPFLFPPPPNQPSP 202
Query: 59 GSLERSPENSPAPEQQPAVTQEQ 81
P P P QP ++ Q
Sbjct: 201 SPSPPPPPPLPPPPPQPPQSRHQ 133
Score = 35.4 bits (80), Expect = 0.14
Identities = 21/60 (35%), Positives = 26/60 (43%)
Frame = -3
Query: 12 PMRQRISPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAP 71
P + PR P + P+R SPP PP PPPP SPS + R +P P
Sbjct: 409 PRSPTVIDPRPPAASAVPLPLRPLYPLRDSPPRRPP-PPPPHSPSILPPSSRLSPPTPLP 233
Score = 34.7 bits (78), Expect = 0.25
Identities = 28/87 (32%), Positives = 30/87 (34%), Gaps = 23/87 (26%)
Frame = -2
Query: 12 PMRQRISPPRQPHRVVLDSPVRTTGVQSPSPP-------PSPPVPPPPPS-----PSQVG 59
P R PP SP R G +P PP P PP PPPPP P Q+
Sbjct: 437 PPTLRPPPPAPVAHGHRPSPPRRIGSSTPPPPFVPIAGFPPPPSPPPPPPLPLHPPPQLS 258
Query: 60 SLE-----------RSPENSPAPEQQP 75
SL SP P P P
Sbjct: 257 SLPPHAPSSFLLPLTSPPPPPPPPHPP 177
Score = 33.9 bits (76), Expect = 0.42
Identities = 24/75 (32%), Positives = 29/75 (38%), Gaps = 9/75 (12%)
Frame = -1
Query: 9 HHSPMRQRISPPRQPHRVVLDSPV-RTTGVQSPSPPP--------SPPVPPPPPSPSQVG 59
HH P +PP L+ P T PS PP SPP PP PP+
Sbjct: 570 HHHP-----NPPASQSPATLNRPTSHTPACPPPSRPPPHPRAPSLSPPPPPHPPTSPPRP 406
Query: 60 SLERSPENSPAPEQQ 74
RS +P P +Q
Sbjct: 405 GRPRSSTLAPPPHRQ 361
Score = 32.3 bits (72), Expect = 1.2
Identities = 21/66 (31%), Positives = 29/66 (43%), Gaps = 6/66 (9%)
Frame = -3
Query: 12 PMRQRISPPRQPHRVV------LDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSP 65
P+ + P QPH + P R + +PPPS +PPPP SP+ + R P
Sbjct: 544 PIPRHPEPTHQPHPCLPPPIPSAPPPTRAFALPPSAPPPSD-LPPPPRSPTVID--PRPP 374
Query: 66 ENSPAP 71
S P
Sbjct: 373 AASAVP 356
Score = 32.0 bits (71), Expect = 1.6
Identities = 24/74 (32%), Positives = 25/74 (33%), Gaps = 16/74 (21%)
Frame = -3
Query: 12 PMRQRISPPRQPHRVVLDSPVRTTGVQSPSPPP----------------SPPVPPPPPSP 55
P R PP P L P R+ V P PP SPP PPPP P
Sbjct: 469 PTRAFALPPSAPPPSDLPPPPRSPTVIDPRPPAASAVPLPLRPLYPLRDSPPRRPPPPPP 290
Query: 56 SQVGSLERSPENSP 69
L S SP
Sbjct: 289 HSPSILPPSSRLSP 248
Score = 30.8 bits (68), Expect = 3.6
Identities = 26/88 (29%), Positives = 29/88 (32%), Gaps = 30/88 (34%)
Frame = -2
Query: 18 SPPRQPHRVVLDSPVRTTGVQSPSPP------------------PSPPVPPPPP-----S 54
SPP +P + P PSPP P PP PPPPP
Sbjct: 452 SPPLRPPTLRPPPPAPVAHGHRPSPPRRIGSSTPPPPFVPIAGFPPPPSPPPPPPLPLHP 273
Query: 55 PSQVGSLERS-------PENSPAPEQQP 75
P Q+ SL P SP P P
Sbjct: 272 PPQLSSLPPHAPSSFLLPLTSPPPPPPP 189
Score = 30.4 bits (67), Expect = 4.6
Identities = 23/78 (29%), Positives = 35/78 (44%), Gaps = 9/78 (11%)
Frame = -1
Query: 7 RQHHSPMRQRISP-------PRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVG 59
+ H SP + P PR P RV+L T+ + + S PP+ P PP+
Sbjct: 708 KAHRSPRPPHVGPSLAHPARPR-PGRVLLVMQYPTSFLGTGSAPPAIHHHPNPPASQSPA 532
Query: 60 SLERSPENSPA--PEQQP 75
+L R ++PA P +P
Sbjct: 531 TLNRPTSHTPACPPPSRP 478
Score = 30.4 bits (67), Expect = 4.6
Identities = 20/63 (31%), Positives = 24/63 (37%)
Frame = -1
Query: 3 TRRRRQHHSPMRQRISPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLE 62
T R H+P PP +P P SP PPP PP PP P + +L
Sbjct: 531 TLNRPTSHTPA---CPPPSRP------PPHPRAPSLSPPPPPHPPTSPPRPGRPRSSTLA 379
Query: 63 RSP 65
P
Sbjct: 378 PPP 370
>TC81043 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproline-rich
glycoprotein DZ-HRGP {Volvox carteri f. nagariensis},
partial (19%)
Length = 555
Score = 43.5 bits (101), Expect = 5e-04
Identities = 25/54 (46%), Positives = 28/54 (51%)
Frame = +1
Query: 18 SPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAP 71
SPP+ P SP +P+PPP PP PP PPSP GS P SPAP
Sbjct: 235 SPPKPPAPPSPGSPP----APAPAPPPIPPSPPAPPSP---GSPPSPPPASPAP 375
Score = 34.7 bits (78), Expect = 0.25
Identities = 17/36 (47%), Positives = 19/36 (52%)
Frame = +1
Query: 40 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 75
P PPSP PP PP+P GS P +PAP P
Sbjct: 211 PPAPPSPGSPPKPPAPPSPGS---PPAPAPAPPPIP 309
Score = 32.7 bits (73), Expect = 0.94
Identities = 16/38 (42%), Positives = 19/38 (49%)
Frame = +1
Query: 39 SPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPA 76
+P P SPP PP PPSP + +P P P PA
Sbjct: 217 APPSPGSPPKPPAPPSPGSPPA--PAPAPPPIPPSPPA 324
Score = 30.8 bits (68), Expect = 3.6
Identities = 15/34 (44%), Positives = 18/34 (52%)
Frame = +1
Query: 42 PPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 75
P P PP PP P SP + + SP + PAP P
Sbjct: 199 PKPPPPAPPSPGSPPKPPA-PPSPGSPPAPAPAP 297
>TC77148 ENOD20
Length = 1108
Score = 42.4 bits (98), Expect = 0.001
Identities = 31/76 (40%), Positives = 36/76 (46%), Gaps = 10/76 (13%)
Frame = +3
Query: 11 SPMRQRISPP-RQPHRVVLDSPVRTTGVQSPSPPPSP-----PVPPP----PPSPSQVGS 60
SP R S P P R L SP + SPSP PSP P+P P P SPS S
Sbjct: 441 SPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASPSPSPS 620
Query: 61 LERSPENSPAPEQQPA 76
L +SP S +P P+
Sbjct: 621 LSKSPSPSESPSLAPS 668
Score = 37.0 bits (84), Expect = 0.050
Identities = 22/55 (40%), Positives = 28/55 (50%), Gaps = 4/55 (7%)
Frame = +3
Query: 26 VVLDSPVRTTGVQSPSPPPSPPVP----PPPPSPSQVGSLERSPENSPAPEQQPA 76
VV+ +PV + SP PPPSPP P P P P + SP SP+P P+
Sbjct: 396 VVMVAPV----LSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPS 548
Score = 30.0 bits (66), Expect = 6.1
Identities = 20/61 (32%), Positives = 27/61 (43%)
Frame = +3
Query: 11 SPMRQRISPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPA 70
+P+ PP P +P+ +S PPSP P P PSPS SP ++P
Sbjct: 408 APVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPS-PSPSPSPSP----SPSPRSTPI 572
Query: 71 P 71
P
Sbjct: 573 P 575
Score = 30.0 bits (66), Expect = 6.1
Identities = 21/61 (34%), Positives = 27/61 (43%), Gaps = 5/61 (8%)
Frame = +3
Query: 5 RRRQHHSP-----MRQRISPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVG 59
R+R SP + + SP P + SP + +PS PS P P PSPS G
Sbjct: 582 RKRSPASPSPSPSLSKSPSPSESPS--LAPSPSDSVASLAPSSSPSDESPSPAPSPSSSG 755
Query: 60 S 60
S
Sbjct: 756 S 758
>TC90865 homologue to GP|9049326|dbj|BAA99336.1 orf129b {Beta vulgaris},
partial (16%)
Length = 940
Score = 42.0 bits (97), Expect = 0.002
Identities = 23/55 (41%), Positives = 33/55 (59%), Gaps = 1/55 (1%)
Frame = -3
Query: 568 NVRRVLLDQGSSADIIYGDAFDKLGLTDNDLTPYAG-TLVGFAGEQVMVRGYIDL 621
+VR+++LD GSS DI+Y + F L L + +L Y G L GF G GY++L
Sbjct: 938 DVRKIILD*GSSCDIMYSNMF*TLQLLEKNLISYVGFDLQGFNGATTRP*GYVEL 774
>TC79965 similar to GP|7549635|gb|AAF63820.1| hypothetical protein
{Arabidopsis thaliana}, partial (15%)
Length = 742
Score = 41.6 bits (96), Expect = 0.002
Identities = 20/35 (57%), Positives = 23/35 (65%), Gaps = 1/35 (2%)
Frame = -3
Query: 38 QSPSPPPSPPVPPPPPS-PSQVGSLERSPENSPAP 71
QSP PPP PP PPPPP PS SP+N+P+P
Sbjct: 371 QSPPPPPPPPKPPPPPKIPS-----PPSPKNNPSP 282
>BG448304
Length = 637
Score = 41.2 bits (95), Expect = 0.003
Identities = 31/117 (26%), Positives = 51/117 (43%), Gaps = 8/117 (6%)
Frame = +2
Query: 399 PEAPKYQPRDANPKKWCEFHRSAGHDTDDCWTLQREIDKLIRAGYQGNRQGLWRNGGDQN 458
P+ P + + + K+C FH+ H TDDC L+ I+ LI+ G N +N
Sbjct: 65 PKTPTQENKGTDKTKYCRFHKCHRHLTDDCIHLKDTIEILIQRGR--------LNQLTKN 220
Query: 459 KAHKREEERADTKGKKKQESAAIATKGADDTFAQH--------SGPPVGTINTIAGG 507
+++ + T K K A++ + D+ F +H + P T N I GG
Sbjct: 221 PEPEKQTVKLITDKKNKDIVVAMSVEQLDE-FVEHVDITPYSCTWEPFPTANVITGG 388
>BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (39%)
Length = 1358
Score = 40.8 bits (94), Expect = 0.003
Identities = 21/39 (53%), Positives = 24/39 (60%)
Frame = -2
Query: 33 RTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAP 71
R TGVQSP P P+ P PP PP P L+R+P PAP
Sbjct: 349 RHTGVQSPVPCPALPHPPGPPPP-----LDRAPPPPPAP 248
Score = 39.3 bits (90), Expect = 0.010
Identities = 26/69 (37%), Positives = 29/69 (41%), Gaps = 3/69 (4%)
Frame = -2
Query: 11 SPMRQRISPPRQPHRVVLDSPVRTTGVQSPSPPPSPP---VPPPPPSPSQVGSLERSPEN 67
S +R R SPP P P + + P PPP PP PPPPP Q P
Sbjct: 235 SRLRPRASPPPPPPP---PGPAASAPPRPPPPPPPPPQRPPPPPPPHTHQ-----PEPPT 80
Query: 68 SPAPEQQPA 76
SP Q PA
Sbjct: 79 SPPRRQPPA 53
Score = 38.5 bits (88), Expect = 0.017
Identities = 24/70 (34%), Positives = 27/70 (38%), Gaps = 3/70 (4%)
Frame = -2
Query: 10 HSPMRQRISPPRQPHRVVLDSPVRTTGVQSPSPPPSP---PVPPPPPSPSQVGSLERSPE 66
H+P ++PPRQ R P PPP P P P PPP P P
Sbjct: 577 HTPAH--LAPPRQSPR--------------PRPPPEPLHNPRPRPPPRPPPAAPARAPPS 446
Query: 67 NSPAPEQQPA 76
PAP PA
Sbjct: 445 GPPAPPTPPA 416
Score = 38.5 bits (88), Expect = 0.017
Identities = 23/64 (35%), Positives = 28/64 (42%), Gaps = 6/64 (9%)
Frame = -2
Query: 18 SPPRQPHRVVLDSPVRTTGVQSPSPPP------SPPVPPPPPSPSQVGSLERSPENSPAP 71
+PP P + L S +R P PPP +PP PPPPP P +R P P
Sbjct: 271 APPPPPAPLPLASRLRPRASPPPPPPPPGPAASAPPRPPPPPPPPP----QRPPPPPPPH 104
Query: 72 EQQP 75
QP
Sbjct: 103 THQP 92
Score = 38.1 bits (87), Expect = 0.022
Identities = 19/46 (41%), Positives = 24/46 (51%)
Frame = -3
Query: 31 PVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPA 76
P R + PP SPP P PP SPS + R+P +P P + PA
Sbjct: 588 PSRYIPPPTSPPPASPPAPVPPQSPSTIRG--RAPPPAPPPRRPPA 457
Score = 38.1 bits (87), Expect = 0.022
Identities = 23/75 (30%), Positives = 32/75 (42%), Gaps = 9/75 (12%)
Frame = -2
Query: 10 HSPMRQRISPPRQPHRVVLDSPVRTTGVQSPSPPP---------SPPVPPPPPSPSQVGS 60
H+ ++ + P PH P+ P+P P SPP PPPPP P+ +
Sbjct: 346 HTGVQSPVPCPALPHPPGPPPPLDRAPPPPPAPLPLASRLRPRASPPPPPPPPGPA-ASA 170
Query: 61 LERSPENSPAPEQQP 75
R P P P Q+P
Sbjct: 169 PPRPPPPPPPPPQRP 125
Score = 37.0 bits (84), Expect = 0.050
Identities = 27/84 (32%), Positives = 32/84 (37%), Gaps = 19/84 (22%)
Frame = -3
Query: 3 TRRRRQHHSPMRQ---------------RISPPRQPHRVVLDSPVRTTGVQSPSPPPS-- 45
T RRR SP Q + PPR R +R + Q PS P
Sbjct: 876 TARRRSSTSPAPQIPPLRRSIVRARAARAVRPPRTAQRHPPRRTLRASAAQHPSRAPRRH 697
Query: 46 --PPVPPPPPSPSQVGSLERSPEN 67
PP PPPP SP +G+ EN
Sbjct: 696 PPPPPPPPPSSPPGMGTRPSCQEN 625
Score = 35.4 bits (80), Expect = 0.14
Identities = 22/60 (36%), Positives = 27/60 (44%)
Frame = -1
Query: 12 PMRQRISPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAP 71
P+ ++I +P V L VR P PP PPVPPPP P E +P P P
Sbjct: 641 PLAKKIGGQPRPALVPL---VRVATYPRPPRPP-PPVPPPPSPPRAPPQSEAAPPPPPPP 474
Score = 33.5 bits (75), Expect = 0.55
Identities = 18/44 (40%), Positives = 20/44 (44%)
Frame = -3
Query: 12 PMRQRISPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSP 55
P R PPR PH P T + P PPP+ PPP P P
Sbjct: 147 PHPPRSGPPR-PH------PPTRTSLSPPPPPPAASPPPPAPPP 37
Score = 33.1 bits (74), Expect = 0.72
Identities = 22/63 (34%), Positives = 26/63 (40%), Gaps = 4/63 (6%)
Frame = -2
Query: 12 PMRQRISPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPP----PPSPSQVGSLERSPEN 67
P R +PP Q H + R T + +PPP P PPP PP L R
Sbjct: 784 PSPHRPAPPPQAH----PARERRTASLARTPPPPPSSPPPAPLLPPRNGNASLLPRKLVG 617
Query: 68 SPA 70
SPA
Sbjct: 616 SPA 608
Score = 31.2 bits (69), Expect = 2.7
Identities = 27/84 (32%), Positives = 33/84 (39%), Gaps = 17/84 (20%)
Frame = -2
Query: 9 HHSPMRQRISP-----------PRQPHRVVLDSPVRTTGVQSPSPPPSPPVPP----PP- 52
H +P RQ P PR P R +P R PS PP+PP PP PP
Sbjct: 565 HLAPPRQSPRPRPPPEPLHNPRPRPPPRPPPAAPARAP----PSGPPAPPTPPATRAPPV 398
Query: 53 -PSPSQVGSLERSPENSPAPEQQP 75
SP G + +P+ Q P
Sbjct: 397 RASPWCPGQGQETPQGRHTGVQSP 326
Score = 30.4 bits (67), Expect = 4.6
Identities = 20/71 (28%), Positives = 24/71 (33%), Gaps = 18/71 (25%)
Frame = -1
Query: 19 PPRQPHRVVLDSPVRTTGV----------------QSPSPPPSPP--VPPPPPSPSQVGS 60
PP P L P G+ +P+PPP+PP PP P P
Sbjct: 269 PPPAPGPTPLSKPPAAEGIPPATAPPPRARRLRPPSAPAPPPTPPAAAPPAPTPPHAPA* 90
Query: 61 LERSPENSPAP 71
P PAP
Sbjct: 89 APHLPPPPPAP 57
Score = 29.6 bits (65), Expect = 7.9
Identities = 15/46 (32%), Positives = 20/46 (42%), Gaps = 5/46 (10%)
Frame = -1
Query: 40 PSPPPSPP-----VPPPPPSPSQVGSLERSPENSPAPEQQPAVTQE 80
PSPP +PP PPPPP P ++P P P ++
Sbjct: 533 PSPPRAPPQSEAAPPPPPPPRGARPRAPLRPPSAPDPAGHPRAPRQ 396
Score = 29.6 bits (65), Expect = 7.9
Identities = 19/51 (37%), Positives = 21/51 (40%), Gaps = 2/51 (3%)
Frame = -2
Query: 11 SPMRQRISPPRQPHRVVLDSPVRTTGVQSPSPPPSPP--VPPPPPSPSQVG 59
+P R PP P R P T P PP SPP PP P +P G
Sbjct: 172 APPRPPPPPPPPPQRPPPPPPPHT---HQPEPPTSPPRRQPPAPRAPPPSG 29
Score = 29.6 bits (65), Expect = 7.9
Identities = 19/60 (31%), Positives = 24/60 (39%), Gaps = 2/60 (3%)
Frame = -3
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPP--PPPSPSQVGSLERSPENSPAPEQQPA 76
PP P V T ++P P P P PP PPP+P + P P + PA
Sbjct: 555 PPASPPAPVPPQSPSTIRGRAPPPAPPPRRPPARPPPAPQR-----------PRPRRPPA 409
>BQ144482 similar to GP|6523547|emb| hydroxyproline-rich glycoprotein DZ-HRGP
{Volvox carteri f. nagariensis}, partial (16%)
Length = 993
Score = 40.4 bits (93), Expect = 0.004
Identities = 19/47 (40%), Positives = 25/47 (52%)
Frame = +3
Query: 18 SPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERS 64
SPP P + +L +R G + P+PPP P P P PS VG R+
Sbjct: 267 SPPPPPPQPLLGPGLRPPGPRPPAPPPPAPAPRPAPSRPSVGRPRRA 407
Score = 36.6 bits (83), Expect = 0.065
Identities = 23/62 (37%), Positives = 27/62 (43%)
Frame = +2
Query: 14 RQRISPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQ 73
+ R +P P +L P + SPS P P PPPPPS S SP PAP
Sbjct: 167 QSRPAPTFSPPPPLLSPPPLPLSLPSPSFAPLPLPPPPPPSASPWP--RPSPPRPPAPRA 340
Query: 74 QP 75
P
Sbjct: 341 PP 346
Score = 36.2 bits (82), Expect = 0.085
Identities = 18/61 (29%), Positives = 24/61 (38%), Gaps = 4/61 (6%)
Frame = +1
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPP----SPPVPPPPPSPSQVGSLERSPENSPAPEQQ 74
PP+ P + P + P PP PP PPPPP P + +P P +
Sbjct: 163 PPKSPRAHLFPPPSPSISAPPPPLPPLPLFRPPPPPPPPPPLSLSLAPAFAPPAPGPPRP 342
Query: 75 P 75
P
Sbjct: 343 P 345
Score = 35.8 bits (81), Expect = 0.11
Identities = 22/67 (32%), Positives = 24/67 (34%)
Frame = +3
Query: 5 RRRQHHSPMRQRISPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERS 64
R R P + PP P P + P PPSP PPPPP P L
Sbjct: 141 RPRDRPPPPKVAPRPPFPPPLPFYLRPPSPSPSPPPLSPPSPSPPPPPPQPLLGPGLRPP 320
Query: 65 PENSPAP 71
PAP
Sbjct: 321 GPRPPAP 341
Score = 34.3 bits (77), Expect = 0.32
Identities = 24/70 (34%), Positives = 29/70 (41%), Gaps = 7/70 (10%)
Frame = +3
Query: 14 RQRISPPRQPHRVVLDSPVRTTGVQSPSPPPSPPV-------PPPPPSPSQVGSLERSPE 66
R R PP+ R P+ ++ PSP PSPP PPPPP +G R P
Sbjct: 147 RDRPPPPKVAPRPPFPPPLPFY-LRPPSPSPSPPPLSPPSPSPPPPPPQPLLGPGLRPPG 323
Query: 67 NSPAPEQQPA 76
P PA
Sbjct: 324 PRPPAPPPPA 353
Score = 32.3 bits (72), Expect = 1.2
Identities = 25/89 (28%), Positives = 31/89 (34%), Gaps = 13/89 (14%)
Frame = +1
Query: 14 RQRISPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVP----------PPPPSPSQVGSLER 63
R + PP P P+ + P PPP PP P PP P P + R
Sbjct: 178 RAHLFPPPSPSISAPPPPLPPLPLFRPPPPPPPPPPLSLSLAPAFAPPAPGPPRPPPPPR 357
Query: 64 SPENSPAPEQ---QPAVTQEQWRHLMRSI 89
P P P + PA WR R +
Sbjct: 358 PPAR-PRPARVLAGPAGRPRHWRASRRRV 441
Score = 31.2 bits (69), Expect = 2.7
Identities = 17/50 (34%), Positives = 23/50 (46%), Gaps = 3/50 (6%)
Frame = +3
Query: 11 SPMRQRISPPRQPHRVVLDSPVRTTGVQSPSPPPS---PPVPPPPPSPSQ 57
SP +SPP P+ G++ P P P PP P P P+PS+
Sbjct: 231 SPSPPPLSPPSPSPPPPPPQPLLGPGLRPPGPRPPAPPPPAPAPRPAPSR 380
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,272,224
Number of Sequences: 36976
Number of extensions: 557529
Number of successful extensions: 17537
Number of sequences better than 10.0: 475
Number of HSP's better than 10.0 without gapping: 5906
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10562
length of query: 1291
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1184
effective length of database: 5,058,295
effective search space: 5989021280
effective search space used: 5989021280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0331c.2


