

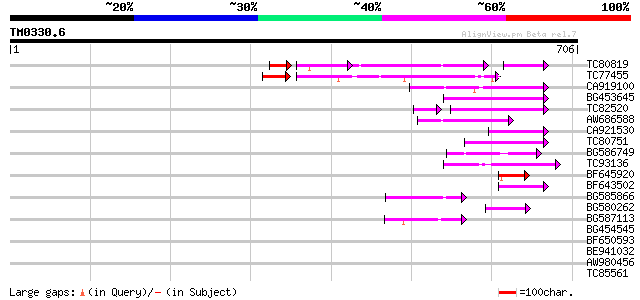
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0330.6
(706 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non... 82 6e-35
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 92 4e-24
CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [i... 89 4e-18
BG453645 72 1e-12
TC82520 55 8e-12
AW686588 66 4e-11
CA921530 59 5e-09
TC80751 weakly similar to GP|13161403|dbj|BAB33036. CPRD49 {Vign... 54 2e-07
BG586749 weakly similar to GP|18138053|emb tropinone reductase I... 53 4e-07
TC93136 46 4e-05
BF645920 45 7e-05
BF643502 44 2e-04
BG585866 44 3e-04
BG580262 42 6e-04
BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At... 42 8e-04
BG454545 38 0.015
BF650593 37 0.020
BE941032 36 0.044
AW980456 36 0.058
TC85561 weakly similar to GP|4530126|gb|AAD21872.1| receptor-lik... 36 0.058
>TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non-LTR
retroelement reverse transcriptase {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 1262
Score = 82.0 bits (201), Expect(4) = 6e-35
Identities = 50/177 (28%), Positives = 82/177 (46%), Gaps = 2/177 (1%)
Frame = +2
Query: 422 SNMGK--WVNGVWQWEFKWRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGG 479
++MG+ W WE W R L E E ++L L +N D WRW L+ G
Sbjct: 380 ADMGRLGWTVDGRAWE--WTRRLFVWEEECVRECCILLNNFVLQDNVNDKWRWLLDPVNG 553
Query: 480 FSFNSSFVFLQDQYLEEPDPVFNWIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVLHN 539
+S + ++ + + +W +PS + FVWR+L R+ T+DNL++ VL
Sbjct: 554 YSVKVFYRYITSTGHISDRSLVDDVWHKHIPSKVSLFVWRLLRNRLPTKDNLVHRGVL-L 730
Query: 540 ALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSDPKVHLLQFSSI 596
A A C C+ ES +HL C+ + WLG+ + + + H +QF+ +
Sbjct: 731 ATNAACVCGCVDSESTTHLFLHCNVFCSLWSLVRNWLGIPSMSSGELRTHFIQFAKM 901
Score = 44.7 bits (104), Expect(4) = 6e-35
Identities = 24/74 (32%), Positives = 35/74 (46%), Gaps = 4/74 (5%)
Frame = +3
Query: 358 SSWWKEIQSITVDNG---WPWFQESVQKTVFAGNATQFWAENWTGSGSLELRFRRLYNLS 414
S WWK I + G WF+E+++ V G FW + W G L L++ RL +L+
Sbjct: 177 SMWWKTICKVREGVGEGVGNWFEENIRMVVGDGRDAFFWYDTWAGDVPLRLKYPRLLDLA 356
Query: 415 CQKREQVS-NMGKW 427
K +V G W
Sbjct: 357 MDKECKVGLTWGGW 398
Score = 41.6 bits (96), Expect(4) = 6e-35
Identities = 16/27 (59%), Positives = 21/27 (77%)
Frame = +1
Query: 324 FNMALLGKWKWRFLTEPDSLWCRVVKA 350
FN++LLGKW WR L + + LW RV+KA
Sbjct: 49 FNLSLLGKWCWRLLVDKEGLWHRVLKA 129
Score = 38.5 bits (88), Expect(4) = 6e-35
Identities = 17/56 (30%), Positives = 31/56 (55%)
Frame = +1
Query: 615 WSIWCLRNMVVFNGGELDKDQLLEQIQVTAWRWLRSRLDDF*YSWYEWKSNPRICL 670
W IW RN +F L+++I++ ++ WL+S+ F YS+ +W N +C+
Sbjct: 946 WVIWKERNNRLFQNTVTAPFVLIDRIKLHSFLWLKSKQVGFAYSYLDWWKNSLLCM 1113
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 91.7 bits (226), Expect(2) = 4e-24
Identities = 77/270 (28%), Positives = 115/270 (42%), Gaps = 16/270 (5%)
Frame = -2
Query: 358 SSWWKEIQSITVDNGWPWFQESVQKTVFAGNATQFWAENWTGSGSLELRF------RRLY 411
S WWK++ S+ WF + + V G ++ FW + W S L F RL
Sbjct: 778 SRWWKDLMSLEEVGRVRWFPRELIRKVGDGRSSFFWKDAWDSSVPLRESFPRAFFPYRLL 599
Query: 412 NLSCQKREQVSNMGKWVNGVWQWEFKWRR-NLQGREVGWFEELMVILQGIRLVENKKDTW 470
+ C ++ G +W WRR L E EL+ L+G+ ++ D W
Sbjct: 598 KMGCGDLWDMNAEGV------RWRLYWRRLELFEWEKERLLELLGRLEGV-VLRYWADIW 440
Query: 471 RWKLEGEGGFSFNSSFVFLQ------DQYLEEPDPVFNWIWMVPVPSNIKAFVWRVLLGR 524
WK + EG FS NS + LQ D+ E + +F +W P+ + AF W + L R
Sbjct: 439 VWKPDKEGVFSVNSCYFLLQNLRLLEDRLSYEEEVIFRELWKSKAPAKVLAFSWTLFLDR 260
Query: 525 IQTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQVS 584
I T NL ++L C C +E+ HL C +* E WL + +S
Sbjct: 259 IPTMVNLGKRRLLRVEDSKRCVFCGCQDETVVHLFLHCDVISKV*REVMRWLNFNL--IS 86
Query: 585 DPKVHLLQFSSIGWS---KAQKLGESAIWM 611
P L +I WS +A+KL + A W+
Sbjct: 85 PPN---LLIHAICWSREVRAKKLRQGA-WL 8
Score = 38.5 bits (88), Expect(2) = 4e-24
Identities = 15/34 (44%), Positives = 23/34 (67%)
Frame = -3
Query: 316 ISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVK 349
+ ++D RL N++LL KW WR L + SLW V++
Sbjct: 945 LGVRDIRLVNVSLLAKWWWRLLQDQSSLWKEVLE 844
>CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [imported] -
Sulfolobus solfataricus, partial (3%)
Length = 789
Score = 89.4 bits (220), Expect = 4e-18
Identities = 57/176 (32%), Positives = 83/176 (46%), Gaps = 4/176 (2%)
Frame = -2
Query: 499 PVFNWIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLAEESGSHL 558
P IW VP + F WR+L R+ T+ NL+ V+ +C C A ES HL
Sbjct: 599 PYAEMIWHRQVPLKVSVFAWRLLRDRLPTKSNLIYRGVIPTEA-GLCVSGCGALESAQHL 423
Query: 559 LFSCSNSMLI*YECHAWLG---VSTAQVSDPKVHLLQF-SSIGWSKAQKLGESAIWMSVL 614
SCS + W+G V T +SD H +QF S G +KA + IW+
Sbjct: 422 FLSCSYFASLWSLVRDWIGFVGVDTNVLSD---HFVQFVHSTGGNKASQSFLQLIWLLCA 252
Query: 615 WSIWCLRNMVVFNGGELDKDQLLEQIQVTAWRWLRSRLDDF*YSWYEWKSNPRICL 670
W +W RN + FN +LL++++ + WL++R F + + W SNP CL
Sbjct: 251 WVLWTERNNMCFNDSITPLPRLLDKVKYLSLGWLKARNASFLFGTFSWWSNPLQCL 84
>BG453645
Length = 622
Score = 71.6 bits (174), Expect = 1e-12
Identities = 36/130 (27%), Positives = 63/130 (47%)
Frame = +3
Query: 541 LEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSDPKVHLLQFSSIGWSK 600
+ + C LC L E+ +H+ C + L+ + W V+ + VH +S G +
Sbjct: 21 VSSTCVLCNLKVETTTHIFLHCDVASLVWFRVMKWFDVNFLIPPNLFVHWECWSEGGSAN 200
Query: 601 AQKLGESAIWMSVLWSIWCLRNMVVFNGGELDKDQLLEQIQVTAWRWLRSRLDDF*YSWY 660
G +W + +W +W RN ++F G + ++E+I+V + RW+ R +Y
Sbjct: 201 RVTKGHWLVWHTTIWVLWAKRNDLIFKGLNCVAEDVIEEIKVLS*RWMLERSSTPSCFFY 380
Query: 661 EWKSNPRICL 670
EW NPR+CL
Sbjct: 381 EWSWNPRLCL 410
>TC82520
Length = 833
Score = 54.7 bits (130), Expect(2) = 8e-12
Identities = 29/123 (23%), Positives = 58/123 (46%), Gaps = 1/123 (0%)
Frame = +2
Query: 549 CLAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSDPKVHLLQFSSIGWS-KAQKLGES 607
C E+ +HL C + WL + +D + +QF+S+ S +
Sbjct: 317 CGDSETATHLFLHCDIFGSLWSHVLRWLHLLLVLPADIRQFFIQFTSMAGSPRFTHSFLQ 496
Query: 608 AIWMSVLWSIWCLRNMVVFNGGELDKDQLLEQIQVTAWRWLRSRLDDF*YSWYEWKSNPR 667
+W + +W +W RN VF D +EQ+++ ++ WL+ + F +S+++W +P
Sbjct: 497 IMWFASVWVLWKKRNNRVFQNSLSDPSTFVEQVKMHSFLWLKFQQATFSFSYHDWWKHPL 676
Query: 668 ICL 670
+C+
Sbjct: 677 LCM 685
Score = 33.9 bits (76), Expect(2) = 8e-12
Identities = 13/34 (38%), Positives = 20/34 (58%)
Frame = +3
Query: 504 IWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVL 537
+W +PS + FVWR+ R+ T+ NL+ VL
Sbjct: 186 VWQKNIPSKVSMFVWRLFHNRLPTKVNLMQRHVL 287
>AW686588
Length = 567
Score = 66.2 bits (160), Expect = 4e-11
Identities = 38/121 (31%), Positives = 62/121 (50%), Gaps = 2/121 (1%)
Frame = +1
Query: 509 VPSNIKAFVWRVLLGRIQTRDNLLN*QVLHNALEAI-CPLCCLAEESGSHLLFSCSNSML 567
VP + WR++ R+ T+ NL+ + L A+EA C + C E+ +HL C+
Sbjct: 142 VPLKVSILAWRLIRDRLPTKANLVRRRCL--AVEAAGCVVGCGIAETANHLFLHCATFGA 315
Query: 568 I*YECHAWLGVSTAQVSDPKVHLLQF-SSIGWSKAQKLGESAIWMSVLWSIWCLRNMVVF 626
+ AW+GVS A D H +QF + G ++A++ IW+ +W +W RN +F
Sbjct: 316 VWQHIRAWIGVSGADPHDLSDHFIQFITCTGHTRARRSFMQLIWLLCVWMVWNERNNRLF 495
Query: 627 N 627
N
Sbjct: 496 N 498
>CA921530
Length = 793
Score = 59.3 bits (142), Expect = 5e-09
Identities = 24/76 (31%), Positives = 45/76 (58%), Gaps = 2/76 (2%)
Frame = -1
Query: 597 GWSKAQKL--GESAIWMSVLWSIWCLRNMVVFNGGELDKDQLLEQIQVTAWRWLRSRLDD 654
GW + +K+ G IW + +W +W N +FN ++ + ++E+++V +WRW +R++
Sbjct: 634 GWERNKKIRRGLWLIWHATIWILWKAWNNRIFNNQAVEAEDIIEEVKVLSWRWTFNRINI 455
Query: 655 F*YSWYEWKSNPRICL 670
+YEW NP+ CL
Sbjct: 454 PVCMYYEWCWNPKYCL 407
>TC80751 weakly similar to GP|13161403|dbj|BAB33036. CPRD49 {Vigna
unguiculata}, partial (52%)
Length = 1156
Score = 54.3 bits (129), Expect = 2e-07
Identities = 31/106 (29%), Positives = 58/106 (54%), Gaps = 2/106 (1%)
Frame = +1
Query: 567 LI*YECHAW-LGVSTAQVSDPKVHLLQFSSIG-WSKAQKLGESAIWMSVLWSIWCLRNMV 624
+I* H W LGVS+ HL+Q G +S++ + +W S +W IW R+ +
Sbjct: 673 II*SFIHKWFLGVSSTDPPCIPDHLIQLVQ*GGYSRSIRSTLLLVWFSYMWVIWKERSKM 852
Query: 625 VFNGGELDKDQLLEQIQVTAWRWLRSRLDDF*YSWYEWKSNPRICL 670
+FN + QLLE++++ ++ WL++ F + ++ + NP +C+
Sbjct: 853 IFNQKKDHVHQLLEKVKLLSF*WLKATNSTFPFDYHSRRLNPLLCM 990
>BG586749 weakly similar to GP|18138053|emb tropinone reductase I {Calystegia
sepium}, partial (11%)
Length = 787
Score = 53.1 bits (126), Expect = 4e-07
Identities = 34/118 (28%), Positives = 53/118 (44%)
Frame = -2
Query: 545 CPLCCLAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSDPKVHLLQFSSIGWSKAQKL 604
C LC E HL C+ + + + WLG D H ++ + +K +
Sbjct: 531 CVLCEGVPEVSEHLFLQCATAK-VWADLLRWLGFDYHTPPDLFFHWKWWNEMSGNKKTRK 355
Query: 605 GESAIWMSVLWSIWCLRNMVVFNGGELDKDQLLEQIQVTAWRWLRSRLDDF*YSWYEW 662
G IW +RN ++FN D+L+E I+V AWRW+ +RL+ +YEW
Sbjct: 354 GY*IIWR--------VRNDLIFNNSRSGMDELVEAIKVLAWRWVLNRLNLPACMFYEW 205
>TC93136
Length = 722
Score = 46.2 bits (108), Expect = 4e-05
Identities = 38/146 (26%), Positives = 62/146 (42%), Gaps = 1/146 (0%)
Frame = +1
Query: 541 LEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSDPKVHLLQFSSIGWSK 600
+E I E+ SHL C + + WL S LL+ +
Sbjct: 226 IEQIDRFAMFYNETSSHLFLHCPFLSSVWSKILGWLYYSKFVCV---FRLLE------RR 378
Query: 601 AQKLGESAIWMSVLWSIWCLRNMVVFNGGELDKDQLLEQIQVTAWRWLRSRLDDF*YSWY 660
++ G IW + +W IW N +F D+++E+I+V +WRW +RL +Y
Sbjct: 379 SRTKGVWLIWHATIWVIWKGINNRIFKNISKAIDEIVEEIKVLSWRWSLTRLKCPPCFYY 558
Query: 661 EWKSNPRICLLS-I*SVGCHGVDCWV 685
EW +P C+ S+G G+ W+
Sbjct: 559 EWCWDPGDCVSR*FLSMGFSGLLPWL 636
>BF645920
Length = 631
Score = 45.4 bits (106), Expect = 7e-05
Identities = 15/42 (35%), Positives = 30/42 (70%), Gaps = 3/42 (7%)
Frame = -3
Query: 609 IWM---SVLWSIWCLRNMVVFNGGELDKDQLLEQIQVTAWRW 647
+W+ + +W+IW RN ++FN G ++ D ++E+++V +WRW
Sbjct: 626 LWLISTTTVWAIWRARNNIIFNEGIVEVDSMVEEVKVLSWRW 501
>BF643502
Length = 631
Score = 44.3 bits (103), Expect = 2e-04
Identities = 20/62 (32%), Positives = 36/62 (57%)
Frame = -1
Query: 609 IWMSVLWSIWCLRNMVVFNGGELDKDQLLEQIQVTAWRWLRSRLDDF*YSWYEWKSNPRI 668
IW+ +W +W RN V+FN LL++I++ + WL+++ F + + +SNP +
Sbjct: 490 IWLLCVWVVWNERNNVMFNHVVTPIPCLLDKIKLLSLGWLKAKNATFVFGIQQGRSNPLV 311
Query: 669 CL 670
CL
Sbjct: 310 CL 305
>BG585866
Length = 828
Score = 43.5 bits (101), Expect = 3e-04
Identities = 29/103 (28%), Positives = 49/103 (47%), Gaps = 2/103 (1%)
Frame = +3
Query: 468 DTWRWKLEGEGGFSFNS--SFVFLQDQYLEEPDPVFNWIWMVPVPSNIKAFVWRVLLGRI 525
D + W G ++ S S++ Q + + + ++WIW + +P K F+W +
Sbjct: 348 DAYIWPHNSNGVYTAKSGYSWILSQTETVNYNNSSWSWIWRLKIPEKYKFFLWLACHNAV 527
Query: 526 QTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI 568
T +LLN + + N+ AIC C EES H + C S +I
Sbjct: 528 PTL-SLLNHRNMVNS--AICSRCGEHEESFFHCVRDCRFSKII 647
>BG580262
Length = 424
Score = 42.4 bits (98), Expect = 6e-04
Identities = 18/56 (32%), Positives = 33/56 (58%)
Frame = +2
Query: 593 FSSIGWSKAQKLGESAIWMSVLWSIWCLRNMVVFNGGELDKDQLLEQIQVTAWRWL 648
FS++ WSK + IW + IW +RN ++F ++ + ++E I++T+ RWL
Sbjct: 173 FSTLSWSKKHPTELTFIWYVTIRKIWEVRNKIIFFLRRVEMEDIVEGIKLTS*RWL 340
>BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (10%)
Length = 767
Score = 42.0 bits (97), Expect = 8e-04
Identities = 24/112 (21%), Positives = 50/112 (44%), Gaps = 10/112 (8%)
Frame = -3
Query: 467 KDTWRWKLEGEGGFSFNSSFVFL--------QDQYLEEP--DPVFNWIWMVPVPSNIKAF 516
+D++ W+ G +S S + Q +++P D ++ +W ++ F
Sbjct: 429 EDSYSWEYSKSGHYSVKSGYYVQTNIIAAANQRGTVDQPSLDDLYQRVWKYNTSPKVRHF 250
Query: 517 VWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI 568
+WR + + T N+ + H + + C C + E+ +H+LF C + LI
Sbjct: 249 LWRCISNSLPTAANMRS---RHISKDGSCSRCGMESETVNHILFQCPYARLI 103
>BG454545
Length = 520
Score = 37.7 bits (86), Expect = 0.015
Identities = 20/60 (33%), Positives = 32/60 (53%)
Frame = +2
Query: 612 SVLWSIWCLRNMVVFNGGELDKDQLLEQIQVTAWRWLRSRLDDF*YSWYEWKSNPRICLL 671
+ +W I RN +F D +++++I+V AW+W RL+ + EW NPR LL
Sbjct: 113 AAIWVI*NTRNHRIFKNVVRDVYEIMDEIKVLAWKWSLVRLNIPTCLYNEWSWNPRDFLL 292
>BF650593
Length = 486
Score = 37.4 bits (85), Expect = 0.020
Identities = 21/54 (38%), Positives = 25/54 (45%)
Frame = +3
Query: 509 VPSNIKAFVWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSC 562
VP + VWR+ + TRDNL VL + C C EES SH F C
Sbjct: 306 VPLKVSCLVWRLFQNXLATRDNLSKRGVL-DQNSIXCVXDCGREESVSHFFFEC 464
>BE941032
Length = 435
Score = 36.2 bits (82), Expect = 0.044
Identities = 29/85 (34%), Positives = 39/85 (45%), Gaps = 3/85 (3%)
Frame = +2
Query: 516 FVWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAW 575
F+W VLL R T+DNLL V+ +A+ C C +HL C+ I W
Sbjct: 152 FLWCVLLNRFPTKDNLLKRGVI-SAIYQSCVGECGNLYDATHLFLHCNFFRQIWINVSDW 328
Query: 576 LG---VSTAQVSDPKVHLLQFSSIG 597
L V+ ++SD L QF S G
Sbjct: 329 LSFVMVTLLRISD---RLAQFGSFG 394
>AW980456
Length = 779
Score = 35.8 bits (81), Expect = 0.058
Identities = 22/67 (32%), Positives = 32/67 (46%), Gaps = 7/67 (10%)
Frame = -2
Query: 472 WKLEGEGGFSFNSSFVFLQDQ-----YLEEPDPVFNW--IWMVPVPSNIKAFVWRVLLGR 524
WK E G + S++ F ++ YL P NW IW + VP ++ VWR+ G
Sbjct: 196 WKDEKHGKYYVKSAYRFCVEELFDSSYLHRPG---NWSGIWKLKVPPKVQNLVWRMCRGC 26
Query: 525 IQTRDNL 531
+ TR L
Sbjct: 25 LPTRIRL 5
>TC85561 weakly similar to GP|4530126|gb|AAD21872.1| receptor-like protein
kinase homolog RK20-1 {Phaseolus vulgaris}, partial (18%)
Length = 1549
Score = 35.8 bits (81), Expect = 0.058
Identities = 18/52 (34%), Positives = 25/52 (47%)
Frame = +2
Query: 618 WCLRNMVVFNGGELDKDQLLEQIQVTAWRWLRSRLDDF*YSWYEWKSNPRIC 669
W RN +FN KDQLL I+ + WL+ F + +Y W S +C
Sbjct: 1259 WKERNSRIFNDNMSSKDQLLASIKFHS*WWLKMHKLGFAFDYYYWWSMASLC 1414
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.345 0.151 0.582
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,439,239
Number of Sequences: 36976
Number of extensions: 573897
Number of successful extensions: 7116
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 3886
Number of HSP's successfully gapped in prelim test: 349
Number of HSP's that attempted gapping in prelim test: 2884
Number of HSP's gapped (non-prelim): 4697
length of query: 706
length of database: 9,014,727
effective HSP length: 103
effective length of query: 603
effective length of database: 5,206,199
effective search space: 3139337997
effective search space used: 3139337997
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0330.6


