

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
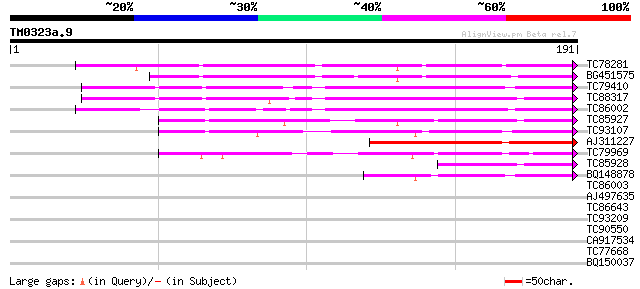
Query= TM0323a.9
(191 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78281 weakly similar to GP|14423500|gb|AAK62432.1 Unknown prot... 129 5e-31
BG451575 similar to SP|Q41495|ST14 STS14 protein precursor. [Pot... 109 6e-25
TC79410 similar to PIR|H84518|H84518 pathogenesis-related PR-1-l... 83 7e-17
TC88317 weakly similar to PIR|T04989|T04989 pathogenesis-related... 74 3e-14
TC86002 weakly similar to PIR|S10205|S10205 pathogenesis-related... 72 1e-13
TC85927 PR-1 71 3e-13
TC93107 weakly similar to GP|13560653|gb|AAK30143.1 pathogenesis... 67 3e-12
AJ311227 similar to PIR|H84518|H845 pathogenesis-related PR-1-li... 66 9e-12
TC79969 weakly similar to PIR|T04232|T04232 pathogenesis-related... 62 1e-10
TC85928 SP|Q40374|PR1_MEDTR Pathogenesis-related protein PR-1 pr... 52 1e-07
BQ148878 weakly similar to GP|4928711|gb|A PR1a precursor {Glyci... 51 2e-07
TC86003 weakly similar to GP|7407641|gb|AAF62171.1| pathogenesis... 34 0.040
AJ497635 similar to GP|688429|dbj|B tumor-related protein {Nicot... 25 0.37
TC86643 similar to GP|5922612|dbj|BAA84613.1 EST AU078118(E3904)... 28 1.7
TC93209 similar to PIR|T51447|T51447 transcription regulator-lik... 27 3.7
TC90550 weakly similar to GP|17065498|gb|AAL32903.1 tyrosine ami... 27 4.8
CA917534 27 4.8
TC77668 similar to GP|18767124|gb|AAL79277.1 unknown {Saccharomy... 27 6.3
BQ150037 26 8.3
>TC78281 weakly similar to GP|14423500|gb|AAK62432.1 Unknown protein
{Arabidopsis thaliana}, partial (71%)
Length = 760
Score = 129 bits (325), Expect = 5e-31
Identities = 77/182 (42%), Positives = 100/182 (54%), Gaps = 13/182 (7%)
Frame = +3
Query: 23 PFFAVLVALALLLVSSQAT------------PATTDGAKVYLFEHNFVRREKGISEHLEW 70
P F L L+LLLVSS A P T AK +L HN R E G+ E L+W
Sbjct: 66 PLFFSLATLSLLLVSSTAARPAATTPEPTAPPPLTPAAKEFLESHNKARAEVGV-EPLQW 242
Query: 71 SDKLAKDASLYARFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDR- 129
S+KLAKD SL R++R+K C +YG NQ + + P + + +WVK++
Sbjct: 243 SEKLAKDTSLLVRYQRNKMACDFANLTASKYGG--NQLWAGSAAAVTPSKAVEEWVKEKE 416
Query: 130 FYYEGNNSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGE 189
FY NN+CV N+ CG YTQ+VW+K+ LGC+QA C L +CFY PPGNV GE
Sbjct: 417 FYIHVNNTCV-VNHECGVYTQVVWKKSAQLGCSQATC-TGKKEASLTICFYDPPGNVIGE 590
Query: 190 RP 191
P
Sbjct: 591 SP 596
>BG451575 similar to SP|Q41495|ST14 STS14 protein precursor. [Potato]
{Solanum tuberosum}, partial (48%)
Length = 427
Score = 109 bits (273), Expect = 6e-25
Identities = 61/145 (42%), Positives = 83/145 (57%), Gaps = 1/145 (0%)
Frame = +1
Query: 48 AKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRDKYLCTPGIFDKKRYGVCCNQ 107
A+ +L HN R G+ E L WS++LA S R++RDK C +YG NQ
Sbjct: 4 AREFLQTHNQARASVGV-EPLTWSEQLANTTSKLVRYQRDKLSCQFANLTAGKYGA--NQ 174
Query: 108 WLEDTVKIMDPREVMRQWVKDR-FYYEGNNSCVPANYPCGDYTQIVWRKTKYLGCAQAEC 166
+ + PR V+ +WVK++ FY +N+CV N+ CG YTQ+VWRK+ LGCAQ C
Sbjct: 175 LMARGAAVT-PRMVVEEWVKEKEFYNHSDNTCV-VNHRCGVYTQVVWRKSVELGCAQTTC 348
Query: 167 GPDYGGVRLNVCFYYPPGNVPGERP 191
G + L++CFYYPPGN G P
Sbjct: 349 GKE--DTXLSICFYYPPGNYVGXSP 417
>TC79410 similar to PIR|H84518|H84518 pathogenesis-related PR-1-like protein
[imported] - Arabidopsis thaliana, partial (84%)
Length = 1002
Score = 82.8 bits (203), Expect = 7e-17
Identities = 52/167 (31%), Positives = 80/167 (47%)
Frame = +2
Query: 25 FAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARF 84
F+++ L L ++ A A Y+ HN RR+ G++ ++ W + LA A YA
Sbjct: 41 FSLICVLGLFIIIGHVAHAQNSQAD-YVNSHNEARRQVGVA-NVVWDNNLATVAQNYANS 214
Query: 85 RRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPANYP 144
RR T RYG + L + + + +R WV ++ Y N++ +
Sbjct: 215 RRGDCRLT---HSGGRYG----ENLAGSTGDLSGTDAVRLWVNEKNDYNYNSNTCASGKV 373
Query: 145 CGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
CG YTQ+VWR TK +GCA+ C G +C Y PPGN G++P
Sbjct: 374 CGHYTQVVWRNTKRIGCAKVRCN---NGGTFIICNYDPPGNYVGQKP 505
>TC88317 weakly similar to PIR|T04989|T04989 pathogenesis-related protein 1
precursor 19.3K - Arabidopsis thaliana, partial (84%)
Length = 658
Score = 74.3 bits (181), Expect = 3e-14
Identities = 52/168 (30%), Positives = 83/168 (48%), Gaps = 1/168 (0%)
Frame = +3
Query: 25 FAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARF 84
F++L L L L+ A + Y+ HN RR+ G++ ++ W + +A A YA
Sbjct: 48 FSLLCVLGLSLIMVHVAHAQNSQSD-YVNAHNDARRQVGVA-NIVWDNTVASFAQDYANQ 221
Query: 85 RR-DKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPANY 143
R+ D L G RYG + L + M + ++ WV ++ Y+ N++ +
Sbjct: 222 RKGDCQLIHSG--GGGRYG----ENLAWSSGDMSGSDAVKLWVNEKADYDYNSNTCASGK 383
Query: 144 PCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
CG YTQ+VWR ++ +GCA+ C + G C Y PPGN GE+P
Sbjct: 384 VCGHYTQVVWRNSQRVGCAKVRCDNNRG--TFITCNYDPPGNYVGEKP 521
>TC86002 weakly similar to PIR|S10205|S10205 pathogenesis-related protein 1
- common tobacco, partial (72%)
Length = 830
Score = 72.4 bits (176), Expect = 1e-13
Identities = 51/169 (30%), Positives = 77/169 (45%)
Frame = +1
Query: 23 PFFAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYA 82
P A+L+A + +Q +P YL HN R + G+ + W K+A A Y
Sbjct: 67 PLMAILLATLTQISYAQNSPQD------YLKIHNKARSDVGVGP-ISWDAKVASYAETYV 225
Query: 83 RFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPAN 142
+ K C + K YG + L + M + W+ ++ YY N++
Sbjct: 226 N--KLKANCKM-VHSKGPYG----ENLAWSSGDMTGTAAVTMWIGEKKYYNYNSNSCAVG 384
Query: 143 YPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
Y CG YTQ+VWR + +GCA+ +C G + C Y PPGN G+RP
Sbjct: 385 YQCGHYTQVVWRDSVRVGCAKVKCND--GRSTIISCNYDPPGNYIGQRP 525
>TC85927 PR-1
Length = 949
Score = 70.9 bits (172), Expect = 3e-13
Identities = 49/144 (34%), Positives = 64/144 (44%), Gaps = 3/144 (2%)
Frame = +2
Query: 51 YLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRDKYLC--TPGIFDKKRYGVCCNQW 108
+L N R G+ L W DKL A YA RR+ + G + + + W
Sbjct: 167 FLIPQNIARAAVGLRP-LVWDDKLTHYAQWYANQRRNDCALEHSNGPYGENIFWGSGVGW 343
Query: 109 LEDTVKIMDPREVMRQWVKDR-FYYEGNNSCVPANYPCGDYTQIVWRKTKYLGCAQAECG 167
+P + + WV ++ FY +NSCV CG YTQ+VW T +GCA C
Sbjct: 344 --------NPAQAVSAWVDEKQFYNYWHNSCVDGEM-CGHYTQVVWGSTTKVGCASVVCS 496
Query: 168 PDYGGVRLNVCFYYPPGNVPGERP 191
D G C Y PPGN GERP
Sbjct: 497 DDKG--TFMTCNYDPPGNYYGERP 562
>TC93107 weakly similar to GP|13560653|gb|AAK30143.1 pathogenesis-related
protein PR-1 precursor {Capsicum annuum}, partial (68%)
Length = 560
Score = 67.4 bits (163), Expect = 3e-12
Identities = 48/144 (33%), Positives = 72/144 (49%), Gaps = 3/144 (2%)
Frame = +1
Query: 51 YLFEHNFVRREKGISEHLEWSDKLAKDASLYA--RFRRDKYLCTPGIFDKKRYGVCCNQW 108
+L HN R E G+ L W L A YA R + + + G + +
Sbjct: 103 FLEVHNQARDEVGVGP-LYWEQTLEAYAQNYANKRIKNCELEHSMGPYGEN--------- 252
Query: 109 LEDTVKIMDPREVMRQWVKDRFYYEGN-NSCVPANYPCGDYTQIVWRKTKYLGCAQAECG 167
L + ++ + ++ W+ ++ Y+ N NSCV N CG YTQI+WR + +LGCA+++C
Sbjct: 253 LAEGYGEVNGTDSVKFWLSEKPNYDYNSNSCV--NDECGHYTQIIWRDSVHLGCAKSKC- 423
Query: 168 PDYGGVRLNVCFYYPPGNVPGERP 191
G +C Y PPGNV GERP
Sbjct: 424 --KNGWVFVICSYSPPGNVEGERP 489
>AJ311227 similar to PIR|H84518|H845 pathogenesis-related PR-1-like protein
[imported] - Arabidopsis thaliana, partial (52%)
Length = 379
Score = 65.9 bits (159), Expect = 9e-12
Identities = 28/70 (40%), Positives = 42/70 (60%)
Frame = +2
Query: 122 MRQWVKDRFYYEGNNSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYY 181
+ W+ ++ YY N++ A Y CG YTQ+VWR + +GCA+ +C + G + C Y
Sbjct: 83 VNMWIGEKRYYNYNSNSCAAGYQCGHYTQVVWRNSVRVGCAKVKC--NNGRSTIISCNYD 256
Query: 182 PPGNVPGERP 191
PPGN G+RP
Sbjct: 257 PPGNYNGQRP 286
>TC79969 weakly similar to PIR|T04232|T04232 pathogenesis-related protein
homolog F14M19.60 - Arabidopsis thaliana, partial (78%)
Length = 1035
Score = 62.4 bits (150), Expect = 1e-10
Identities = 47/152 (30%), Positives = 65/152 (41%), Gaps = 11/152 (7%)
Frame = +2
Query: 51 YLFEHNFVRREKG---------ISEHLEW-SDKLAKDASLYARFRRDKYLCTPGIFDKKR 100
+LF HN VR K + ++ W + + D + F D + I
Sbjct: 263 FLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENI----- 427
Query: 101 YGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEG-NNSCVPANYPCGDYTQIVWRKTKYL 159
Y + W P + ++ W + YY NSCV CG YTQIVW+ T+ +
Sbjct: 428 YWGSGSDWT--------PTDAVKAWADEEKYYTYVTNSCVSGQM-CGHYTQIVWKSTRRI 580
Query: 160 GCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
GCA+ C D G V + C Y P GN GERP
Sbjct: 581 GCARVVC--DDGDVFM-TCNYDPVGNYVGERP 667
>TC85928 SP|Q40374|PR1_MEDTR Pathogenesis-related protein PR-1 precursor.
[Barrel medic] {Medicago truncatula}, partial (92%)
Length = 795
Score = 52.4 bits (124), Expect = 1e-07
Identities = 24/47 (51%), Positives = 26/47 (55%)
Frame = +1
Query: 145 CGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
CG YTQ+VW T +GCA C D G C Y PPGN GERP
Sbjct: 361 CGHYTQVVWGSTTKVGCASVVCSDDKG--TFMTCNYDPPGNYYGERP 495
>BQ148878 weakly similar to GP|4928711|gb|A PR1a precursor {Glycine max},
partial (44%)
Length = 450
Score = 51.2 bits (121), Expect = 2e-07
Identities = 23/73 (31%), Positives = 38/73 (51%), Gaps = 1/73 (1%)
Frame = +2
Query: 120 EVMRQWVKDRFYYEGN-NSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVC 178
+ ++ W ++ +++ N C C +TQ+VW + LGC + +C G C
Sbjct: 116 QAVKGWADEKPHFDNYLNKCFDGE--CHHFTQVVWSGSLRLGCGKVKCN---NGGTFVTC 280
Query: 179 FYYPPGNVPGERP 191
YYPPGN+PG+ P
Sbjct: 281 NYYPPGNIPGQLP 319
>TC86003 weakly similar to GP|7407641|gb|AAF62171.1| pathogenesis-related
protein 1 {Betula pendula}, partial (95%)
Length = 356
Score = 33.9 bits (76), Expect = 0.040
Identities = 27/99 (27%), Positives = 41/99 (41%)
Frame = +1
Query: 51 YLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRDKYLCTPGIFDKKRYGVCCNQWLE 110
YL HN R + G+ + W K+A A Y + K C + K YG + L
Sbjct: 61 YLKIHNKARSDVGVGP-ISWDAKVASYAETYVN--KLKANCKM-VHSKGPYG----ENLA 216
Query: 111 DTVKIMDPREVMRQWVKDRFYYEGNNSCVPANYPCGDYT 149
+ M + W+ ++ YY N++ Y CG YT
Sbjct: 217 WSSGDMTGTAAVTMWIGEKKYYNYNSNSCAVGYQCGHYT 333
>AJ497635 similar to GP|688429|dbj|B tumor-related protein {Nicotiana glauca
x Nicotiana langsdorffii}, partial (22%)
Length = 445
Score = 25.4 bits (54), Expect(2) = 0.37
Identities = 8/12 (66%), Positives = 10/12 (82%)
Frame = +1
Query: 180 YYPPGNVPGERP 191
YYPPGN G++P
Sbjct: 103 YYPPGNYVGDKP 138
Score = 23.9 bits (50), Expect(2) = 0.37
Identities = 8/13 (61%), Positives = 9/13 (68%)
Frame = +2
Query: 145 CGDYTQIVWRKTK 157
C YTQ+VW TK
Sbjct: 5 CRHYTQVVWSNTK 43
>TC86643 similar to GP|5922612|dbj|BAA84613.1 EST AU078118(E3904) corresponds
to a region of the predicted gene.~similar to Arabidopsis
thaliana, partial (39%)
Length = 1705
Score = 28.5 bits (62), Expect = 1.7
Identities = 12/39 (30%), Positives = 22/39 (55%)
Frame = -1
Query: 145 CGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPP 183
C + ++W +T++LG + AEC P + L + F+ P
Sbjct: 1102 CRNMLHMIWEQTEHLGESTAECVPSWSHHILPIEFHGLP 986
>TC93209 similar to PIR|T51447|T51447 transcription regulator-like protein -
Arabidopsis thaliana, partial (10%)
Length = 721
Score = 27.3 bits (59), Expect = 3.7
Identities = 12/33 (36%), Positives = 16/33 (48%)
Frame = +3
Query: 52 LFEHNFVRREKGISEHLEWSDKLAKDASLYARF 84
L+E +E S ++W K A LYARF
Sbjct: 471 LYERILSAKENSTSAEIKWKTKDASSTDLYARF 569
>TC90550 weakly similar to GP|17065498|gb|AAL32903.1 tyrosine
aminotransferase-like protein {Arabidopsis thaliana},
partial (44%)
Length = 925
Score = 26.9 bits (58), Expect = 4.8
Identities = 15/39 (38%), Positives = 23/39 (58%)
Frame = +1
Query: 42 PATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASL 80
P +GA + E NF KGI + +++ DKLAK+ S+
Sbjct: 259 PHKPEGAMSAMVEINFAH-VKGIIDDVDFCDKLAKEESV 372
>CA917534
Length = 616
Score = 26.9 bits (58), Expect = 4.8
Identities = 10/24 (41%), Positives = 13/24 (53%)
Frame = +1
Query: 104 CCNQWLEDTVKIMDPREVMRQWVK 127
CC QW +K + R +RQ VK
Sbjct: 85 CCEQWKNKYLKAQESRNALRQAVK 156
>TC77668 similar to GP|18767124|gb|AAL79277.1 unknown {Saccharomyces
cerevisiae}, partial (26%)
Length = 362
Score = 26.6 bits (57), Expect = 6.3
Identities = 13/41 (31%), Positives = 21/41 (50%), Gaps = 1/41 (2%)
Frame = -2
Query: 119 REVMRQWVKDRFYYEGNNSCVPAN-YPCGDYTQIVWRKTKY 158
R V R+ +K Y ++C+ N Y C Q++W KT +
Sbjct: 277 RRVKREKLKF*IYLVPRSNCILKNRYRC*GLVQVLWNKTSW 155
>BQ150037
Length = 628
Score = 26.2 bits (56), Expect = 8.3
Identities = 12/35 (34%), Positives = 16/35 (45%), Gaps = 6/35 (17%)
Frame = -1
Query: 91 CTPGIFDKKRYG------VCCNQWLEDTVKIMDPR 119
C G +YG CC+ W +T+KI D R
Sbjct: 481 CNTGELSLSKYGEKHLRNFCCSLWGGETIKIFDVR 377
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.142 0.477
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,866,318
Number of Sequences: 36976
Number of extensions: 128166
Number of successful extensions: 691
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 674
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 675
length of query: 191
length of database: 9,014,727
effective HSP length: 91
effective length of query: 100
effective length of database: 5,649,911
effective search space: 564991100
effective search space used: 564991100
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0323a.9


