

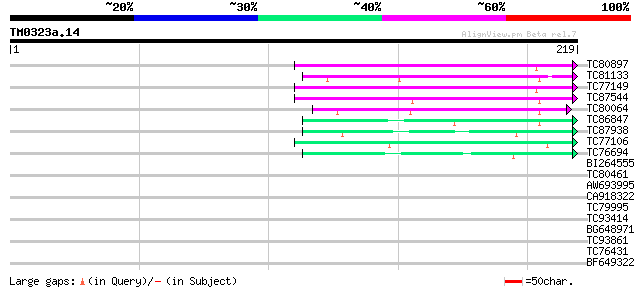
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0323a.14
(219 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80897 similar to GP|3241854|dbj|BAA28872.1 thaumatin-like prot... 71 3e-13
TC81133 similar to PIR|T05493|T05493 pathogenesis-related protei... 67 4e-12
TC77149 similar to GP|3643249|gb|AAC36740.1| thaumatin-like prot... 65 3e-11
TC87544 similar to SP|P50699|TLPH_ARATH Thaumatin-like protein p... 60 9e-10
TC80064 similar to PIR|G86333|G86333 hypothetical protein AAF799... 58 2e-09
TC86847 similar to GP|7406716|emb|CAB85637.1 putative thaumatin-... 49 2e-06
TC87938 similar to GP|20385169|gb|AAM21199.1 pathogenesis-relate... 44 4e-05
TC77106 weakly similar to SP|Q41350|OLP1_LYCES Osmotin-like prot... 44 5e-05
TC76694 homologue to GP|3549691|emb|CAA09228.1 thaumatin-like pr... 42 2e-04
BI264555 similar to GP|3643249|gb| thaumatin-like protein precur... 39 0.002
TC80461 similar to GP|21592749|gb|AAM64698.1 putative thaumatin-... 33 0.11
AW693995 similar to PIR|T05606|T056 protein kinase homolog F9D16... 28 2.7
CA918322 similar to GP|14517466|gb AT3g52150/F4F15_260 {Arabidop... 28 2.7
TC79995 similar to GP|10178093|dbj|BAB11480. gene_id:MVP2.2~pir|... 28 3.6
TC93414 homologue to GP|17907114|emb|CAD13249. tyrosine kinase {... 27 6.1
BG648971 similar to GP|20514808|gb| Putative NBS-LRR type resist... 27 6.1
TC93861 similar to GP|15451102|gb|AAK96822.1 Unknown protein {Ar... 27 6.1
TC76431 similar to GP|8132441|gb|AAF73291.1| extensin {Pisum sat... 27 8.0
BF649322 similar to GP|15021754|gb| root nodule extensin {Pisum ... 27 8.0
>TC80897 similar to GP|3241854|dbj|BAA28872.1 thaumatin-like protein
precursor {Pyrus pyrifolia}, partial (60%)
Length = 602
Score = 71.2 bits (173), Expect = 3e-13
Identities = 36/111 (32%), Positives = 52/111 (46%), Gaps = 2/111 (1%)
Frame = +1
Query: 111 GYNFNLPVTIYSVDLKCKEIQCLVDMMSFCPD*LAIYDSGGLKVGCKNACYTVGDQKLCC 170
G+N + VT CK C ++ CP L + S G V CK+AC + + K CC
Sbjct: 121 GFNVPMSVTPQGGSGDCKTSSCPGNINVVCPSELQVIGSDGSVVACKSACLALNEDKYCC 300
Query: 171 SAVYAFPKKCELNDYRQLVDDNCPYFISNAFN--QTHFTCYGTASYVISFC 219
Y +KC DY + + CP S A++ + FTC+ Y I+FC
Sbjct: 301 RGDYNTEEKCPPTDYSMVFKNQCPGAYSYAYDDKSSTFTCFARPDYAITFC 453
>TC81133 similar to PIR|T05493|T05493 pathogenesis-related protein 19K4.140
- Arabidopsis thaliana, partial (62%)
Length = 1248
Score = 67.4 bits (163), Expect = 4e-12
Identities = 40/115 (34%), Positives = 55/115 (47%), Gaps = 9/115 (7%)
Frame = +2
Query: 114 FNLPVTIY----SVDLKCKEIQCLVDMMSFCPD*LAIYDS---GGLKVGCKNACYTVGDQ 166
+NLP+ I + C C VD+ + CP L + S G V CK+AC GD
Sbjct: 464 YNLPILIEPHGETGGGNCTATGCFVDLNAACPMELKVISSNNGGEESVACKSACEAFGDP 643
Query: 167 KLCCSAVYAFPKKCELNDYRQLVDDNCPYFISNAFNQ--THFTCYGTASYVISFC 219
+ CCS Y P C+ + Y Q CP S A++ + FTC +A Y I+FC
Sbjct: 644 QYCCSGAYGSPDTCKPSSYSQFFKSACPRAYSYAYDDGTSTFTC-ASADYTITFC 805
>TC77149 similar to GP|3643249|gb|AAC36740.1| thaumatin-like protein
precursor Mdtl1 {Malus x domestica}, partial (88%)
Length = 976
Score = 64.7 bits (156), Expect = 3e-11
Identities = 33/111 (29%), Positives = 51/111 (45%), Gaps = 2/111 (1%)
Frame = +2
Query: 111 GYNFNLPVTIYSVDLKCKEIQCLVDMMSFCPD*LAIYDSGGLKVGCKNACYTVGDQKLCC 170
G+N + VT CK C ++ S CP L + S G V CK+AC + CC
Sbjct: 425 GFNVPMSVTPQGGSGDCKSSSCPANINSVCPAELQLKGSDGSVVACKSACLAFNTDQYCC 604
Query: 171 SAVYAFPKKCELNDYRQLVDDNCPYFISNAFN--QTHFTCYGTASYVISFC 219
+ C +Y ++ ++ CP S A++ + FTC +Y I+FC
Sbjct: 605 RGSFNTEATCPPTNYSEIFENQCPEAYSYAYDDKSSTFTCSNGPNYAITFC 757
>TC87544 similar to SP|P50699|TLPH_ARATH Thaumatin-like protein precursor.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (92%)
Length = 1039
Score = 59.7 bits (143), Expect = 9e-10
Identities = 32/111 (28%), Positives = 51/111 (45%), Gaps = 2/111 (1%)
Frame = +3
Query: 111 GYNFNLPVTIYSVDLKCKEIQCLVDMMSFCPD*LAIYDSGGLKV-GCKNACYTVGDQKLC 169
GYN + +T + KC C+ D+ CP L + V GC++AC + C
Sbjct: 450 GYNLPISITPFKGSGKCSYAGCVSDLNLMCPVGLQVRSRDKKHVVGCRSACAAFNSPRYC 629
Query: 170 CSAVYAFPKKCELNDYRQLVDDNCPYFISNAFNQ-THFTCYGTASYVISFC 219
C+ + P+ C+ Y ++ CP S A++ T ASY+I+FC
Sbjct: 630 CTGNFGTPQTCKPTAYSRIFKTACPKAYSYAYDDPTSIATCTNASYMITFC 782
>TC80064 similar to PIR|G86333|G86333 hypothetical protein AAF79910.1
[imported] - Arabidopsis thaliana, partial (75%)
Length = 898
Score = 58.2 bits (139), Expect = 2e-09
Identities = 33/113 (29%), Positives = 49/113 (43%), Gaps = 13/113 (11%)
Frame = +2
Query: 118 VTIYSVDL----------KCKEIQCLVDMMSFCPD*LAIYDSGGLK-VGCKNACYTVGDQ 166
+T+Y+V + C C+ D+ CP L + G V CK+AC
Sbjct: 524 LTVYNVPMLVAPQGGSGDNCTTTGCISDLNGACPSELRVMSVDGKHGVACKSACEAFNSP 703
Query: 167 KLCCSAVYAFPKKCELNDYRQLVDDNCPYFISNAFNQ--THFTCYGTASYVIS 217
+ CCS Y P C+ + Y QL + CP S A++ + FTC Y I+
Sbjct: 704 EYCCSGAYGTPDTCKPSTYSQLFKNACPRAYSYAYDDKTSTFTCANAVDYTIT 862
>TC86847 similar to GP|7406716|emb|CAB85637.1 putative thaumatin-like
protein {Vitis vinifera}, partial (81%)
Length = 960
Score = 48.9 bits (115), Expect = 2e-06
Identities = 31/114 (27%), Positives = 44/114 (38%), Gaps = 8/114 (7%)
Frame = +2
Query: 114 FNLPVTIYSVDLKCKEIQCLVDMMSFCPD*LAIYDSGGLKVGCKNACYTVGDQKLCC--- 170
FN+P+ + C + C D+ CP L + GC N C T + CC
Sbjct: 380 FNIPIQLKPSLNSCGTVNCTADINGECPTPLKV------PGGCNNPCTTFDTTEYCCNSD 541
Query: 171 ---SAVYAFPKKCELNDYRQLVDDNCPYFISNAFNQ--THFTCYGTASYVISFC 219
SA C Y + D CPY S + + F+C G +Y + FC
Sbjct: 542 SARSAATNASATCGPTTYSKFFKDRCPYAYSYPKDDATSTFSCMGGTTYDVVFC 703
>TC87938 similar to GP|20385169|gb|AAM21199.1 pathogenesis-related protein
5-1 {Helianthus annuus}, partial (93%)
Length = 882
Score = 44.3 bits (103), Expect = 4e-05
Identities = 29/109 (26%), Positives = 44/109 (39%), Gaps = 3/109 (2%)
Frame = +1
Query: 114 FNLPVTIYSVDLKC-KEIQCLVDMMSFCPD*LAIYDSGGLKVGCKNACYTVGDQKLCCSA 172
FN+P+ C + I+C D++ CP+ L GC NAC CC++
Sbjct: 388 FNVPMEFSPTSNGCTRGIRCTADIIGQCPNELKTQG------GCNNACTVFKTDNYCCNS 549
Query: 173 VYAFPKKCELNDYRQLVDDNCP--YFISNAFNQTHFTCYGTASYVISFC 219
C +Y Q + CP Y + F+C G +Y + FC
Sbjct: 550 -----GNCGPTNYSQYFKNKCPDAYSYPKDDATSTFSCKGGTNYKVVFC 681
>TC77106 weakly similar to SP|Q41350|OLP1_LYCES Osmotin-like protein
precursor. [Tomato] {Lycopersicon esculentum}, partial
(88%)
Length = 1071
Score = 43.9 bits (102), Expect = 5e-05
Identities = 28/113 (24%), Positives = 45/113 (39%), Gaps = 4/113 (3%)
Frame = +3
Query: 111 GYNFNLPVTIYSVDLKCKEIQCLVDMMSFCPD*LA--IYDSGGLKVGCKNACYTVGDQKL 168
G+N L VT + +C + C D+++ CP L + G V CK+ C +
Sbjct: 435 GFNTPLTVTPHEGKGECPVVGCKADLVASCPPVLQHRVPMGHGPVVACKSGCEAFHSDEH 614
Query: 169 CCSAVYAFPKKCELNDYRQLVDDNCPYFISNAFNQTHF--TCYGTASYVISFC 219
CC + P+ C+ Y + D CP + A + C + FC
Sbjct: 615 CCRNHFNNPQTCKPTVYSKFFKDACPATFTFAHDSPSLIHQCSSPGELKVIFC 773
>TC76694 homologue to GP|3549691|emb|CAA09228.1 thaumatin-like protein PR-5b
{Cicer arietinum}, complete
Length = 947
Score = 42.0 bits (97), Expect = 2e-04
Identities = 26/108 (24%), Positives = 43/108 (39%), Gaps = 2/108 (1%)
Frame = +1
Query: 114 FNLPVTIYSVDLKCKEIQCLVDMMSFCPD*LAIYDSGGLKVGCKNACYTVGDQKLCCSAV 173
FN+P+ + ++ C +I C D+ CP+ L + GC N C + CC+
Sbjct: 409 FNIPMDFFPLNGGCHKISCTADINGQCPNELR------TQGGCNNPCTVFKTNEYCCTNG 570
Query: 174 YAFPKKCELNDYRQLVDDNC--PYFISNAFNQTHFTCYGTASYVISFC 219
C + + D C Y + FTC ++Y + FC
Sbjct: 571 QG---SCGPTKFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFC 705
>BI264555 similar to GP|3643249|gb| thaumatin-like protein precursor Mdtl1
{Malus x domestica}, partial (77%)
Length = 659
Score = 38.5 bits (88), Expect = 0.002
Identities = 21/74 (28%), Positives = 32/74 (42%), Gaps = 2/74 (2%)
Frame = +2
Query: 114 FNLPVTIYSVDLK--CKEIQCLVDMMSFCPD*LAIYDSGGLKVGCKNACYTVGDQKLCCS 171
FN+P++I CK C ++ CP L + G V CK+AC + C+
Sbjct: 437 FNVPMSIVPQGGTGDCKPSSCPANINDVCPTELQMKGPDGKVVACKSACAAFNXPQYXCT 616
Query: 172 AVYAFPKKCELNDY 185
+ P KC+ Y
Sbjct: 617 GEFNSPDKCKPTQY 658
>TC80461 similar to GP|21592749|gb|AAM64698.1 putative thaumatin-like
protein {Arabidopsis thaliana}, partial (63%)
Length = 983
Score = 32.7 bits (73), Expect = 0.11
Identities = 18/61 (29%), Positives = 24/61 (38%)
Frame = +2
Query: 111 GYNFNLPVTIYSVDLKCKEIQCLVDMMSFCPD*LAIYDSGGLKVGCKNACYTVGDQKLCC 170
GYN + V C C D+ CP L + D G C +AC G + CC
Sbjct: 572 GYNLPMLVATSGGTGPCDVTGCSSDLNKKCPSELRVDDGG----ACNSACGAFGKPEYCC 739
Query: 171 S 171
+
Sbjct: 740 N 742
>AW693995 similar to PIR|T05606|T056 protein kinase homolog F9D16.210 -
Arabidopsis thaliana, partial (14%)
Length = 644
Score = 28.1 bits (61), Expect = 2.7
Identities = 14/33 (42%), Positives = 20/33 (60%), Gaps = 1/33 (3%)
Frame = -2
Query: 109 RHGYNFNLPVTIYS-VDLKCKEIQCLVDMMSFC 140
+ G+NF + IY+ + +KCKEI C SFC
Sbjct: 298 QQGHNF---LAIYTTICIKCKEISCFSTSSSFC 209
>CA918322 similar to GP|14517466|gb AT3g52150/F4F15_260 {Arabidopsis
thaliana}, partial (52%)
Length = 662
Score = 28.1 bits (61), Expect = 2.7
Identities = 15/30 (50%), Positives = 18/30 (60%)
Frame = +2
Query: 14 RSSGPILIPSQDTQLFPPESSSSPKTSILL 43
R+ ILIP QDT+L P S S + ILL
Sbjct: 215 RAKSSILIPKQDTRLKEPFSDSISRRLILL 304
>TC79995 similar to GP|10178093|dbj|BAB11480.
gene_id:MVP2.2~pir||T01176~similar to unknown protein
{Arabidopsis thaliana}, partial (48%)
Length = 1028
Score = 27.7 bits (60), Expect = 3.6
Identities = 10/26 (38%), Positives = 16/26 (61%)
Frame = +3
Query: 115 NLPVTIYSVDLKCKEIQCLVDMMSFC 140
NLP+ ++SVDL +E ++ FC
Sbjct: 375 NLPLILFSVDLHVREADLQAQLLEFC 452
>TC93414 homologue to GP|17907114|emb|CAD13249. tyrosine kinase {Schistosoma
mansoni}, partial (1%)
Length = 578
Score = 26.9 bits (58), Expect = 6.1
Identities = 12/29 (41%), Positives = 15/29 (51%)
Frame = +1
Query: 23 SQDTQLFPPESSSSPKTSILLMSPFMVGH 51
S T LFPP + +LL+SPF H
Sbjct: 271 SHPTSLFPPSADPLQTPPLLLLSPFHHSH 357
>BG648971 similar to GP|20514808|gb| Putative NBS-LRR type resistance protein
{Oryza sativa (japonica cultivar-group)}, partial (2%)
Length = 830
Score = 26.9 bits (58), Expect = 6.1
Identities = 15/48 (31%), Positives = 22/48 (45%), Gaps = 1/48 (2%)
Frame = +1
Query: 170 CSAVYAFPKKC-ELNDYRQLVDDNCPYFISNAFNQTHFTCYGTASYVI 216
C FPK+ +L + R +V +NC +S F TC T + I
Sbjct: 511 CPYFSHFPKQLTQLQELRHIVIENCFSLVSTPFRIGELTCLKTLTVFI 654
>TC93861 similar to GP|15451102|gb|AAK96822.1 Unknown protein {Arabidopsis
thaliana}, partial (35%)
Length = 913
Score = 26.9 bits (58), Expect = 6.1
Identities = 13/45 (28%), Positives = 23/45 (50%), Gaps = 5/45 (11%)
Frame = -1
Query: 121 YSVDLKCKE-----IQCLVDMMSFCPD*LAIYDSGGLKVGCKNAC 160
Y L+C+ + CLV FC D +A+++S + + N+C
Sbjct: 604 YRASLQCQPYTNIMVPCLVVCHPFCSDTMALHNSSSVHIPEVNSC 470
>TC76431 similar to GP|8132441|gb|AAF73291.1| extensin {Pisum sativum},
partial (30%)
Length = 890
Score = 26.6 bits (57), Expect = 8.0
Identities = 14/39 (35%), Positives = 22/39 (55%)
Frame = -2
Query: 125 LKCKEIQCLVDMMSFCPD*LAIYDSGGLKVGCKNACYTV 163
L+CKE + +V+ MS C L + +SG G ++A V
Sbjct: 343 LECKEGELVVEGMSICMASLVVVESGSRA*G*RDALVVV 227
>BF649322 similar to GP|15021754|gb| root nodule extensin {Pisum sativum},
partial (54%)
Length = 476
Score = 26.6 bits (57), Expect = 8.0
Identities = 14/39 (35%), Positives = 22/39 (55%)
Frame = -2
Query: 125 LKCKEIQCLVDMMSFCPD*LAIYDSGGLKVGCKNACYTV 163
L+CKE + +V+ MS C L + +SG G ++A V
Sbjct: 364 LECKEGELVVEGMSICMASLVVVESGSRA*G*RDALVVV 248
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.342 0.150 0.506
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,059,737
Number of Sequences: 36976
Number of extensions: 126118
Number of successful extensions: 944
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 931
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 934
length of query: 219
length of database: 9,014,727
effective HSP length: 92
effective length of query: 127
effective length of database: 5,612,935
effective search space: 712842745
effective search space used: 712842745
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0323a.14


