

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
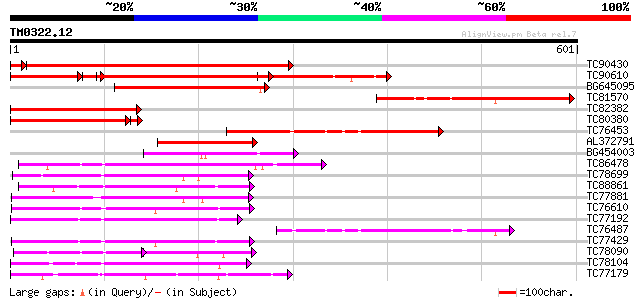
Query= TM0322.12
(601 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC90430 similar to GP|18766642|gb|AAL79042.1 NIMA-related protei... 536 e-157
TC90610 similar to GP|18766642|gb|AAL79042.1 NIMA-related protei... 315 e-129
BG645095 similar to GP|9294413|dbj kinase-like protein {Arabidop... 227 9e-60
TC81570 similar to GP|18766642|gb|AAL79042.1 NIMA-related protei... 211 8e-55
TC82382 homologue to GP|18766642|gb|AAL79042.1 NIMA-related prot... 193 2e-49
TC80380 similar to PIR|T49136|T49136 protein kinase-like protein... 187 1e-48
TC76453 similar to GP|18766642|gb|AAL79042.1 NIMA-related protei... 180 1e-45
AL372791 homologue to GP|15290125|db putative kinase {Oryza sati... 167 1e-41
BG454003 homologue to GP|15290125|db putative kinase {Oryza sati... 155 5e-38
TC86478 similar to GP|3688193|emb|CAA08995.1 MAP3K alpha 1 prote... 132 3e-31
TC78699 weakly similar to PIR|G96761|G96761 probable MAP kinase ... 118 7e-27
TC88861 weakly similar to GP|17027283|gb|AAL34137.1 putative pro... 115 6e-26
TC77881 homologue to GP|15528439|emb|CAC69137. MEK map kinase ki... 114 1e-25
TC76610 similar to PIR|T50802|T50802 serine/threonine protein ki... 114 1e-25
TC77192 similar to SP|P42818|KPK1_ARATH Serine/threonine-protein... 113 2e-25
TC76487 similar to GP|18766642|gb|AAL79042.1 NIMA-related protei... 108 6e-24
TC77429 similar to GP|15215664|gb|AAK91377.1 AT5g25110/T11H3_120... 107 1e-23
TC78090 GP|15528441|emb|CAC69138. MAP kinase kinase {Medicago sa... 72 2e-23
TC78104 homologue to GP|19568098|gb|AAL89456.1 osmotic stress-ac... 106 2e-23
TC77179 similar to GP|2665890|gb|AAB88537.1| calcium-dependent p... 106 2e-23
>TC90430 similar to GP|18766642|gb|AAL79042.1 NIMA-related protein kinase
{Populus x canescens}, partial (45%)
Length = 976
Score = 536 bits (1382), Expect(2) = e-157
Identities = 258/282 (91%), Positives = 271/282 (95%)
Frame = +1
Query: 19 LLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPFIVEYKDSWVEKGCFVC 78
LLVRHKHEKK YVLKKIRLARQ+DRTRRSAHQEMELISKVRNPFIVEYKDSWVEKGCFVC
Sbjct: 130 LLVRHKHEKKKYVLKKIRLARQTDRTRRSAHQEMELISKVRNPFIVEYKDSWVEKGCFVC 309
Query: 79 IIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVNHILHRDVKCSNIFLT 138
II+GYCE GD+A+ +KKANGVNFPEEKLCKWLVQLLMALDYLHVNHILHRDVKCSNIFLT
Sbjct: 310 IIIGYCEGGDMAETVKKANGVNFPEEKLCKWLVQLLMALDYLHVNHILHRDVKCSNIFLT 489
Query: 139 KNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPYGSKSDIWSLGCCIYEMAA 198
KNQDIRLGDFGLAK+LTSDDLASSIVGTPSYMCPELLADIPYGSKSDIWSLGCC+YEMAA
Sbjct: 490 KNQDIRLGDFGLAKLLTSDDLASSIVGTPSYMCPELLADIPYGSKSDIWSLGCCVYEMAA 669
Query: 199 LKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLRKNPELRPPASELLNHPHL 258
+PAFKAFDIQALI+KINKSIV+PLPTMYSSAFRGLVKSMLRKNPELRP A ELLNHPHL
Sbjct: 670 HRPAFKAFDIQALIHKINKSIVSPLPTMYSSAFRGLVKSMLRKNPELRPTAGELLNHPHL 849
Query: 259 QPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVST 300
QPYILKIH KLNSPRRS FP QWP+SNY RRTRF+ P+SVST
Sbjct: 850 QPYILKIHQKLNSPRRSAFPLQWPDSNYGRRTRFMEPESVST 975
Score = 36.2 bits (82), Expect(2) = e-157
Identities = 16/18 (88%), Positives = 17/18 (93%)
Frame = +3
Query: 1 MEQYEVLEQIGKGSFGSA 18
MEQYEVLEQIGKG+FG A
Sbjct: 75 MEQYEVLEQIGKGAFGCA 128
>TC90610 similar to GP|18766642|gb|AAL79042.1 NIMA-related protein kinase
{Populus x canescens}, partial (46%)
Length = 1507
Score = 315 bits (806), Expect(3) = e-129
Identities = 152/187 (81%), Positives = 166/187 (88%)
Frame = +2
Query: 93 IKKANGVNFPEEKLCKWLVQLLMALDYLHVNHILHRDVKCSNIFLTKNQDIRLGDFGLAK 152
+K+ V+F EE+LCKWLVQLLMALDYLH NHILHRDVKCSNIFLTK+QDIRLGDFGLAK
Sbjct: 536 LKRRTVVHFSEERLCKWLVQLLMALDYLHANHILHRDVKCSNIFLTKDQDIRLGDFGLAK 715
Query: 153 MLTSDDLASSIVGTPSYMCPELLADIPYGSKSDIWSLGCCIYEMAALKPAFKAFDIQALI 212
MLTSDDLASSIVGTPSYMCPELLADIPYGSKSDIWSLGCC+YEMAA KPAFKA D+QALI
Sbjct: 716 MLTSDDLASSIVGTPSYMCPELLADIPYGSKSDIWSLGCCVYEMAAHKPAFKALDMQALI 895
Query: 213 NKINKSIVAPLPTMYSSAFRGLVKSMLRKNPELRPPASELLNHPHLQPYILKIHLKLNSP 272
NKINKS+VAPLPTMYS FR +VKSMLRKNPELRP A++LLNHPHLQPYI + P
Sbjct: 896 NKINKSLVAPLPTMYSGTFRSMVKSMLRKNPELRPSAADLLNHPHLQPYIP*SSSETK*P 1075
Query: 273 RRSTFPF 279
++ FPF
Sbjct: 1076QKKYFPF 1096
Score = 147 bits (372), Expect(3) = e-129
Identities = 73/77 (94%), Positives = 73/77 (94%)
Frame = +3
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
MEQYEVLEQIGKGSF SALLVRHKHE K YVLKKIRLARQSDR RRSAHQEMELISKVRN
Sbjct: 258 MEQYEVLEQIGKGSFASALLVRHKHENKRYVLKKIRLARQSDRIRRSAHQEMELISKVRN 437
Query: 61 PFIVEYKDSWVEKGCFV 77
PFIVEYKDSWVEKGCFV
Sbjct: 438 PFIVEYKDSWVEKGCFV 488
Score = 139 bits (351), Expect = 2e-33
Identities = 80/145 (55%), Positives = 100/145 (68%), Gaps = 3/145 (2%)
Frame = +3
Query: 263 LKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVSTLSDQDKCLSLSNDRSLNPSISG 322
LK+HLKLN+PRRSTFPFQW +SN+ RR+RF+ +SVST+S + K LS SNDR+LNPSISG
Sbjct: 1047 LKVHLKLNNPRRSTFPFQWTDSNHARRSRFLERESVSTISGRAKRLSFSNDRALNPSISG 1226
Query: 323 TEQDSQCFTQKAHGLSTSSKEVYEVSVGGACGPWNTNK---TKSPTVERTPRSRADKDST 379
TE S C TQ+A G ST SK YE+S+G N N TK V++ R A K+S
Sbjct: 1227 TEVGSLCSTQRAQGFSTCSKH-YELSIGCVREEHNANNLKDTKFSIVDQMQRLGACKESA 1403
Query: 380 AARRQTMGPSKISRSGSKHDTLPVS 404
RRQT PSKI+ + SK D+L +S
Sbjct: 1404 IPRRQTT-PSKIAYTSSKRDSLIIS 1475
Score = 41.2 bits (95), Expect(3) = e-129
Identities = 16/24 (66%), Positives = 21/24 (86%)
Frame = +1
Query: 78 CIIVGYCERGDLADAIKKANGVNF 101
CI++GYCE GD+A+AIKKAN +F
Sbjct: 490 CIVIGYCEGGDMAEAIKKANCCSF 561
>BG645095 similar to GP|9294413|dbj kinase-like protein {Arabidopsis
thaliana}, partial (35%)
Length = 805
Score = 227 bits (579), Expect = 9e-60
Identities = 108/175 (61%), Positives = 139/175 (78%), Gaps = 11/175 (6%)
Frame = +2
Query: 112 QLLMALDYLHVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMC 171
QLL+A+DYLH N +LHRD+KCSNIFLTK +IRLGDFGLAK+L +DD ASS+VGT +YMC
Sbjct: 5 QLLLAVDYLHSNRVLHRDLKCSNIFLTKENNIRLGDFGLAKLLDTDDPASSVVGTLNYMC 184
Query: 172 PELLADIPYGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAF 231
PE+ +D+PYG KSDIWSLGCC++E+ A +PAF+A D LINKIN+S ++PLP +YSS
Sbjct: 185 PEIFSDMPYGYKSDIWSLGCCMFEIVAHQPAFRAPDRAGLINKINRSTISPLPIVYSSTL 364
Query: 232 RGLVKSMLRKNPELRPPASELLNHPHLQPYILK----------IHL-KLNSPRRS 275
+ ++KSMLRKNPE RP A+ELL HPHLQP++L+ +HL NSP++S
Sbjct: 365 KQIIKSMLRKNPEHRPTAAELLRHPHLQPFVLRSRDAPSVFLPVHLISCNSPKKS 529
>TC81570 similar to GP|18766642|gb|AAL79042.1 NIMA-related protein kinase
{Populus x canescens}, partial (20%)
Length = 895
Score = 211 bits (536), Expect = 8e-55
Identities = 137/223 (61%), Positives = 154/223 (68%), Gaps = 14/223 (6%)
Frame = +3
Query: 390 KISRSGSKHDTLPVSHAPSVKFPPPTRRASHPLPTRASMAKATLYTNVGTFPRVDS-DVS 448
KI +G K ++LPV APS K PTRRAS PL TR + YTNV DS +VS
Sbjct: 6 KIHVTGPKRESLPVPRAPSGKSAMPTRRASLPLHTRGRNT-TSFYTNVDY---ADSPNVS 173
Query: 449 VNAPQIDKIAEFPLASCEDLPFFPVHVPSSTSVHCYSS-SAGSAYRSITKEKC--IHEDK 505
V+APQIDK+AEF AS ED PFF V SST+ +SS S GSA +ITK+KC + + K
Sbjct: 174 VDAPQIDKMAEFSTASYED-PFFHVIRRSSTASAKHSSTSTGSADCTITKDKCTILVDKK 350
Query: 506 VTVPGGSC--------PKGS--ECSKHVTAGVSIHSSAELHQRRFDTSSYQQRAEALEGL 555
VTVP PKGS ECS +VT GVS SS+E Q RFDTSSYQQRAEALEGL
Sbjct: 351 VTVPTSITDAGTDVRFPKGSASECSNYVTTGVSSRSSSESRQHRFDTSSYQQRAEALEGL 530
Query: 556 LEFSARLLQHQRFDELGVLLKPFGLEKVSPRETAIWLAKSFKE 598
LEFS+RLLQ RFDELGVLLKPFG EKVSPRETAIWLAKSFK+
Sbjct: 531 LEFSSRLLQQHRFDELGVLLKPFGPEKVSPRETAIWLAKSFKQ 659
>TC82382 homologue to GP|18766642|gb|AAL79042.1 NIMA-related protein kinase
{Populus x canescens}, partial (22%)
Length = 714
Score = 193 bits (490), Expect = 2e-49
Identities = 99/140 (70%), Positives = 109/140 (77%), Gaps = 1/140 (0%)
Frame = +1
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
MEQYEVLEQIGKG+FGSALLVRHKHEKK YVLKKIRLARQ++R+RRSAHQEMELISK+RN
Sbjct: 286 MEQYEVLEQIGKGAFGSALLVRHKHEKKKYVLKKIRLARQTERSRRSAHQEMELISKLRN 465
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGV-NFPEEKLCKWLVQLLMALDY 119
PFIVEYKDSWVEKGC+VCII+GYCE + K+ V FP L L L
Sbjct: 466 PFIVEYKDSWVEKGCYVCIIIGYCEGWRYGGSYKRRLMVFCFPRRNFASGLFNFLWHLTT 645
Query: 120 LHVNHILHRDVKCSNIFLTK 139
NHILHRDVKCSNIFL +
Sbjct: 646 CT*NHILHRDVKCSNIFLDR 705
>TC80380 similar to PIR|T49136|T49136 protein kinase-like protein -
Arabidopsis thaliana, partial (14%)
Length = 791
Score = 187 bits (476), Expect(2) = 1e-48
Identities = 85/128 (66%), Positives = 109/128 (84%)
Frame = +3
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
ME+YEV+EQIG+G+FG+A LV HK EKK YVLKKIRLA+Q+++ +R+AHQEM LI+K+ N
Sbjct: 363 MEEYEVIEQIGRGAFGAAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLNN 542
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
+IV+Y+D+WVEK VCII GYCE GD+AD+IKKA G FPEEK+CKWL QLL+A+DYL
Sbjct: 543 SYIVDYRDAWVEKEDHVCIITGYCEGGDMADSIKKARGSFFPEEKVCKWLTQLLIAVDYL 722
Query: 121 HVNHILHR 128
H N ++HR
Sbjct: 723 HSNRVIHR 746
Score = 24.3 bits (51), Expect(2) = 1e-48
Identities = 9/12 (75%), Positives = 12/12 (100%)
Frame = +1
Query: 129 DVKCSNIFLTKN 140
D+KCS+IFLTK+
Sbjct: 748 DLKCSHIFLTKD 783
>TC76453 similar to GP|18766642|gb|AAL79042.1 NIMA-related protein kinase
{Populus x canescens}, partial (13%)
Length = 673
Score = 180 bits (457), Expect = 1e-45
Identities = 110/234 (47%), Positives = 142/234 (60%), Gaps = 3/234 (1%)
Frame = +3
Query: 230 AFRGLVKSMLRKNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRR 289
+FR LVKSMLRKNPELRP ASELL HPHLQPY+LKIHLKLNSPRR+T P W ES Y ++
Sbjct: 3 SFRSLVKSMLRKNPELRPSASELLGHPHLQPYVLKIHLKLNSPRRNTLPAHWQESKYTKK 182
Query: 290 TRFVVPDSVSTLSDQDKCLSLSNDRSLNPSISGTEQDSQCFTQKAHGLSTS--SKEVYEV 347
TRF+VP+ + +DK S SNDR+LNPS+SG +QDS C T + G + ++ + E+
Sbjct: 183 TRFLVPED----TVRDKRYSYSNDRTLNPSVSGADQDSVCSTLEI-GCTPDHLNQRLAEL 347
Query: 348 SVGGACGPWNTNKTKSPTVERTPRSRADKDSTAARRQTMGPSKISRSGSKHDTLPVSHAP 407
G + + P V RT S K + G ++ S S + LPVSH+
Sbjct: 348 CTGDS---RDMKLVHKPVVSRT--SSIAKAPKFTSSKVSGTNRKSMESSNNRKLPVSHS- 509
Query: 408 SVKFPPPTRRASHPLPTRASMAKATLYTNVGTFPRVDS-DVSVNAPQIDKIAEF 460
K RRAS PLPTR + + +VG V S D+S N+P+ID+IAEF
Sbjct: 510 ITKSAHTNRRASFPLPTRGGIREPPSRKSVGLLSHVSSPDISDNSPRIDRIAEF 671
>AL372791 homologue to GP|15290125|db putative kinase {Oryza sativa (japonica
cultivar-group)}, partial (11%)
Length = 398
Score = 167 bits (423), Expect = 1e-41
Identities = 76/106 (71%), Positives = 91/106 (85%)
Frame = +3
Query: 157 DDLASSIVGTPSYMCPELLADIPYGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKIN 216
DDLASS+VGTP+YMCPELLADIPYG KSDIWSLGCC+YEMAA +PAFKAFD+ LI+KIN
Sbjct: 3 DDLASSVVGTPNYMCPELLADIPYGFKSDIWSLGCCVYEMAAHRPAFKAFDMAGLISKIN 182
Query: 217 KSIVAPLPTMYSSAFRGLVKSMLRKNPELRPPASELLNHPHLQPYI 262
+S + PLP YS + + L+K MLRK+PE RP ASE+L HP+LQPY+
Sbjct: 183 RSSIGPLPPCYSPSLKILIKGMLRKSPEHRPTASEVLKHPYLQPYV 320
>BG454003 homologue to GP|15290125|db putative kinase {Oryza sativa (japonica
cultivar-group)}, partial (13%)
Length = 660
Score = 155 bits (391), Expect = 5e-38
Identities = 94/220 (42%), Positives = 116/220 (52%), Gaps = 56/220 (25%)
Frame = +2
Query: 143 IRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPYGSKSDIWSLGCCIYEMAALKP- 201
IRLGDFGLAK L DDLASS+VGTP+YMCPELLADIPYG KSDIWSLGCC+YEMAA +P
Sbjct: 2 IRLGDFGLAKTLKQDDLASSVVGTPNYMCPELLADIPYGXKSDIWSLGCCVYEMAAHRPA 181
Query: 202 --AFKAF----------------------------------------------------- 206
AF +F
Sbjct: 182 FKAFVSFF*PFLLYRQHDECLPCDSLHCFVLDFYVCGDITPCVISLISCVFNNVL*IMLQ 361
Query: 207 DIQALINKINKSIVAPLPTMYSSAFRGLVKSMLRKNPELRPPASELLNHPHLQPYILKIH 266
D+ LI+KIN+S + PLP YS + + L+K MLRK+PE RP ASE+L HP+LQPY+
Sbjct: 362 DMAGLISKINRSSIGPLPPCYSPSLKILIKGMLRKSPEHRPTASEVLKHPYLQPYV--DQ 535
Query: 267 LKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVSTLSDQDK 306
+ + + F + P S+ R + ST S DK
Sbjct: 536 YRPSFAPLTAFSPEKPISSATYTKRHMAESXNSTSSSSDK 655
>TC86478 similar to GP|3688193|emb|CAA08995.1 MAP3K alpha 1 protein kinase
{Brassica napus}, partial (52%)
Length = 2266
Score = 132 bits (333), Expect = 3e-31
Identities = 97/350 (27%), Positives = 171/350 (48%), Gaps = 24/350 (6%)
Frame = +2
Query: 10 IGKGSFGSALLVRHKHEKKLYVLKKIRLA---RQSDRTRRSAHQEMELISKVRNPFIVEY 66
+G+G+FG L + ++ +K++R+ + S + HQE++L++++ +P IV+Y
Sbjct: 989 LGRGTFGHVYLGFNSENGQMCAIKEVRVGCDDQNSKECLKQLHQEIDLLNQLSHPNIVQY 1168
Query: 67 KDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVNHIL 126
S + + + + + Y G + +++ F E + + Q++ L YLH + +
Sbjct: 1169 LGSELGEES-LSVYLEYVSGGSIHKLLQEYGP--FKEPVIQNYTRQIVSGLAYLHGRNTV 1339
Query: 127 HRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADI-PYGSKSD 185
HRD+K +NI + N +I+L DFG+AK +TS S G+P +M PE++ + Y D
Sbjct: 1340 HRDIKGANILVDPNGEIKLADFGMAKHITSAASMLSFKGSPYWMAPEVVMNTNGYSLPVD 1519
Query: 186 IWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAP-LPTMYSSAFRGLVKSMLRKNPE 244
IWSLGC + EMAA KP + ++ A I KI S P +P S+ + + L+++P
Sbjct: 1520 IWSLGCTLIEMAASKPPWSQYEGVAAIFKIGNSKDMPIIPEHLSNDAKNFIMLCLQRDPS 1699
Query: 245 LRPPASELLNHPHLQ-------------PYILKIH-----LKLNSPRRSTFPFQWPESNY 286
RP A +LL HP ++ Y+ L+ NS RRS F + +Y
Sbjct: 1700 ARPTAQKLLEHPFIRDQSATKAATRDVSSYMFDGSRTPPVLEPNSNRRSITSF---DGDY 1870
Query: 287 MRRTRFVVPDSVSTLSDQDKCL-SLSNDRSLNPSISGTEQDSQCFTQKAH 335
++ P + + D + + SL S +P ++ CF H
Sbjct: 1871 ATKSAIAAPRTTRSPRDHSRTITSLPVSPSSSPLRQYGQEHRSCFYSPPH 2020
>TC78699 weakly similar to PIR|G96761|G96761 probable MAP kinase T9L24.32
[imported] - Arabidopsis thaliana, partial (71%)
Length = 1119
Score = 118 bits (295), Expect = 7e-27
Identities = 87/266 (32%), Positives = 132/266 (48%), Gaps = 11/266 (4%)
Frame = +3
Query: 4 YEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN-PF 62
+E L +G G+ G+ VRHK +Y LK I TRR A E+ ++ + +
Sbjct: 201 FEKLSVLGHGNGGTVYKVRHKLTSIIYALK-INHYDSDPTTRRRALTEVNILRRATDCTN 377
Query: 63 IVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHV 122
+V+Y S+ + VCI++ Y + G L A+K F E KL +L L YLH
Sbjct: 378 VVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTG--TFSESKLSTVARDILNGLTYLHA 551
Query: 123 NHILHRDVKCSNIFLTKNQDIRLGDFGLAKML-TSDDLASSIVGTPSYMCPELLADIPYG 181
+I HRD+K SNI + ++++ DFG++K + + + +S VGT +YM PE YG
Sbjct: 552 RNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYG 731
Query: 182 S-----KSDIWSLGCCIYEMAA----LKPAFKAFDIQALINKINKSIVAPLPTMYSSAFR 232
+DIWSLG ++E+ + + D +L+ I S LP SS FR
Sbjct: 732 GNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFR 911
Query: 233 GLVKSMLRKNPELRPPASELLNHPHL 258
V+ L+K R A++LL HP L
Sbjct: 912 NFVECCLKKESGERWSAAQLLTHPFL 989
>TC88861 weakly similar to GP|17027283|gb|AAL34137.1 putative protein kinase
{Oryza sativa (japonica cultivar-group)}, partial (34%)
Length = 1847
Score = 115 bits (287), Expect = 6e-26
Identities = 80/261 (30%), Positives = 129/261 (48%), Gaps = 11/261 (4%)
Frame = +1
Query: 10 IGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTR---RSAHQEMELISKVRNPFIVEY 66
IG+GSFGS + LK++ L ++ + QE+ ++ ++ +P IVEY
Sbjct: 1042 IGRGSFGSVYHATNLETGASCALKEVDLVPDDPKSTDCIKQLDQEIRILGQLHHPNIVEY 1221
Query: 67 KDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVNHIL 126
S V G +CI + Y G L ++ GV E + + +L L YLH +
Sbjct: 1222 YGSEVV-GDRLCIYMEYVHPGSLQKFMQDHCGV-MTESVVRNFTRHILSGLAYLHSTKTI 1395
Query: 127 HRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELL-------ADIP 179
HRD+K +N+ + + ++L DFG++K+LT S+ G+P +M PEL+ +
Sbjct: 1396 HRDIKGANLLVDASGIVKLADFGVSKILTEKSYELSLKGSPYWMAPELMMAAMKNETNPT 1575
Query: 180 YGSKSDIWSLGCCIYEMAALKPAFKAF-DIQALINKINKSIVAPLPTMYSSAFRGLVKSM 238
DIWSLGC I EM KP + F QA+ +++S +P S + ++
Sbjct: 1576 VAMAVDIWSLGCTIIEMLTGKPPWSEFPGHQAMFKVLHRS--PDIPKTLSPEGQDFLEQC 1749
Query: 239 LRKNPELRPPASELLNHPHLQ 259
++NP RP A+ LL HP +Q
Sbjct: 1750 FQRNPADRPSAAVLLTHPFVQ 1812
>TC77881 homologue to GP|15528439|emb|CAC69137. MEK map kinase kinsae
{Medicago sativa subsp. x varia}, complete
Length = 1546
Score = 114 bits (285), Expect = 1e-25
Identities = 85/266 (31%), Positives = 133/266 (49%), Gaps = 10/266 (3%)
Frame = +2
Query: 3 QYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPF 62
+ E L +IG GS G+ V H+ + Y LK I + RR H+E++++ V +
Sbjct: 464 ELERLNRIGSGSGGTVYKVVHRINGRAYALKVI-YGHHEESVRRQIHREIQILRDVDDVN 640
Query: 63 IVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEE-KLCKWLVQLLMALDYLH 121
+V+ + + + + +++ Y + G L G + P+E +L Q+L L YLH
Sbjct: 641 VVKCHEMY-DHNAEIQVLLEYMDGGSL-------EGKHIPQENQLADVARQILRGLAYLH 796
Query: 122 VNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSD-DLASSIVGTPSYMCPELL-ADIP 179
HI+HRD+K SN+ + + +++ DFG+ ++L D +S VGT +YM PE + DI
Sbjct: 797 RRHIVHRDIKPSNLLINSRKQVKIADFGVGRILNQTMDPCNSSVGTIAYMSPERINTDIN 976
Query: 180 YGS----KSDIWSLGCCIYEMAALKPAF---KAFDIQALINKINKSIVAPLPTMYSSAFR 232
G DIWSLG I E + F + D +L+ I S PT S FR
Sbjct: 977 DGQYDAYAGDIWSLGVSILEFYMGRFPFAVGRQGDWASLMCAICMSQPPEAPTTASPEFR 1156
Query: 233 GLVKSMLRKNPELRPPASELLNHPHL 258
V L+++P R AS LL+HP L
Sbjct: 1157DFVSRCLQRDPSRRWTASRLLSHPFL 1234
>TC76610 similar to PIR|T50802|T50802 serine/threonine protein kinase-like
protein - Arabidopsis thaliana, partial (73%)
Length = 1799
Score = 114 bits (284), Expect = 1e-25
Identities = 76/263 (28%), Positives = 139/263 (51%), Gaps = 6/263 (2%)
Frame = +2
Query: 3 QYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLAR-QSDRTRRSAHQEMELISKVRNP 61
+YE+ + +G+G+F R+ + +K I+ + + +R + +E+ ++ VR+P
Sbjct: 323 KYEIGKTLGQGNFAKVYHGRNIATNENVAIKVIKKEKLKKERLMKQIKREVSVMRLVRHP 502
Query: 62 FIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLC-KWLVQLLMALDYL 120
IVE K+ K V ++V Y + G+L + K EE + K+ QL+ A+D+
Sbjct: 503 HIVELKEVMANKAK-VFMVVEYVKGGELFAKVAKGK----MEENIARKYFQQLISAVDFC 667
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKM---LTSDDLASSIVGTPSYMCPELLAD 177
H + HRD+K N+ L +N+D+++ DFGL+ + SD + + GTP+Y+ PE+L
Sbjct: 668 HSRGVTHRDLKPENLLLDENEDLKVSDFGLSALPDQRRSDGMLLTPCGTPAYVAPEVLKK 847
Query: 178 IPY-GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVK 236
I Y GSK+DIWS G ++ + F+ ++ + +K K+ LP S + L+
Sbjct: 848 IGYDGSKADIWSCGVILFALLCGYLPFQGENVMRIYSKSFKADYV-LPEWISPGAKKLIS 1024
Query: 237 SMLRKNPELRPPASELLNHPHLQ 259
++L +PE R +++ P Q
Sbjct: 1025NLLVVDPEKRFSIPDIMKDPWFQ 1093
>TC77192 similar to SP|P42818|KPK1_ARATH Serine/threonine-protein kinase
AtPK1/AtPK6 (EC 2.7.1.-). [Mouse-ear cress] {Arabidopsis
thaliana}, partial (76%)
Length = 1992
Score = 113 bits (282), Expect = 2e-25
Identities = 71/247 (28%), Positives = 127/247 (50%), Gaps = 1/247 (0%)
Frame = +3
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQ-EMELISKVR 59
+E +EVL+ +G+G+F VR K ++Y +K +R + ++ + E E+++K+
Sbjct: 759 IEDFEVLKVVGQGAFAKVYQVRKKGTSEIYAMKVMRKDKIMEKNHAEYMKAEREILTKIE 938
Query: 60 NPFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDY 119
+PF+V+ + S+ K + +++ + G L + F E+ + +++ A+ +
Sbjct: 939 HPFVVQLRYSFQTK-YRLYLVLDFVNGGHLFFQLYHQG--LFREDLARIYAAEIVSAVSH 1109
Query: 120 LHVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIP 179
LH I+HRD+K NI + + + L DFGLAK ++S+ GT YM PE++
Sbjct: 1110 LHSKGIMHRDLKPENILMDADGHVMLTDFGLAKKFEESTRSNSLCGTAEYMAPEIILGKG 1289
Query: 180 YGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSML 239
+ +D WS+G ++EM KP F + Q + KI K + LP SS L+K +L
Sbjct: 1290 HDKAADWWSVGILLFEMLTGKPPFCGGNCQKIQQKIVKDKI-KLPGFLSSDAHALLKGLL 1466
Query: 240 RKNPELR 246
K R
Sbjct: 1467 NKEAPKR 1487
>TC76487 similar to GP|18766642|gb|AAL79042.1 NIMA-related protein kinase
{Populus x canescens}, partial (16%)
Length = 852
Score = 108 bits (270), Expect = 6e-24
Identities = 91/274 (33%), Positives = 135/274 (49%), Gaps = 21/274 (7%)
Frame = +2
Query: 283 ESNYMRRTRFVVPDSVSTLSDQDKCLSLSNDRSLNPSISGTEQDSQCFTQKAHGLSTS-- 340
ES Y ++TRF+VP+ +DK S SNDR+LNPS+SG +QDS C T + G +
Sbjct: 77 ESKYTKKTRFLVPEDTV----RDKRYSYSNDRTLNPSVSGADQDSVCSTLEI-GCTPDHL 241
Query: 341 SKEVYEVSVGGACGPWNTNKTKSPTVERTPRSRADKDSTAARRQTMGPSKISRSGSKHDT 400
++ + E+ G + + P V RT S K + G ++ S S +
Sbjct: 242 NQRLAELCTGDS---RDMKLVHKPVVSRT--SSIAKAPKFTSSKVSGTNRKSMESSNNRK 406
Query: 401 LPVSHAPSVKFPPPTRRASHPLPTRASMAKATLYTNVGTFPRVDS-DVSVNAPQIDKIAE 459
LPVSH+ K RRAS PLPTR + + +VG V S D+SVN+P+ID+IAE
Sbjct: 407 LPVSHS-ITKSAHTNRRASCPLPTRGGIREPPSRKSVGLLSHVSSPDISVNSPRIDRIAE 583
Query: 460 FPLASCEDLPFFPVHVPSSTSVHCYSSSAGSAYRSITKEKCIHE--DKVTVPGGSC---- 513
FPL+S +D FPV+ S+++ S+ S +KC E D+ +V S
Sbjct: 584 FPLSSYDDC-LFPVNKMSTSA----QGSSSFPSHSTVIDKCTIEVCDRASVKPSSTDDWQ 748
Query: 514 ------------PKGSECSKHVTAGVSIHSSAEL 535
K ++ TAG S ++S++L
Sbjct: 749 GIKRSMLKEIHEDKSGSSDQNATAGASSYTSSDL 850
>TC77429 similar to GP|15215664|gb|AAK91377.1 AT5g25110/T11H3_120
{Arabidopsis thaliana}, partial (79%)
Length = 1869
Score = 107 bits (267), Expect = 1e-23
Identities = 72/262 (27%), Positives = 131/262 (49%), Gaps = 5/262 (1%)
Frame = +3
Query: 3 QYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLAR-QSDRTRRSAHQEMELISKVRNP 61
+YE+ +GKG+F + + +K + + + + +E+ ++ V++P
Sbjct: 267 KYEMGRVLGKGTFAKVYYAKEISTGEGVAIKVVDKDKVKKEGMMEQIKREISVMRLVKHP 446
Query: 62 FIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLH 121
IV K+ K + ++ Y G+L + + K F EE K+ QL+ A+DY H
Sbjct: 447 NIVNLKEVMATK-TKILFVMEYARGGELFEKVAKGK---FKEELARKYFQQLISAVDYCH 614
Query: 122 VNHILHRDVKCSNIFLTKNQDIRLGDFGLAKM---LTSDDLASSIVGTPSYMCPELLADI 178
+ HRD+K N+ L +N+++++ DFGL+ + L D L + GTP+Y+ PE++
Sbjct: 615 SRGVSHRDLKPENLLLDENENLKVSDFGLSALPEHLRQDGLLHTQCGTPAYVAPEVVRKR 794
Query: 179 PY-GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKS 237
Y G K+D WS G +Y + A F+ ++ ++ NK+ K P +S + L+
Sbjct: 795 GYSGFKADTWSCGVILYALLAGFLPFQHENLISMYNKVFKE-EYQFPPWFSPESKRLISK 971
Query: 238 MLRKNPELRPPASELLNHPHLQ 259
+L +PE R S ++N P Q
Sbjct: 972 ILVADPERRITISSIMNVPWFQ 1037
>TC78090 GP|15528441|emb|CAC69138. MAP kinase kinase {Medicago sativa subsp.
x varia}, complete
Length = 1677
Score = 71.6 bits (174), Expect(2) = 2e-23
Identities = 42/142 (29%), Positives = 80/142 (55%), Gaps = 1/142 (0%)
Frame = +2
Query: 5 EVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPFIV 64
++++ +GKG+ G LV+HK + + LK I++ + + R+ +E+++ + P++V
Sbjct: 431 DIVKVVGKGNGGVVQLVQHKWTNQFFALKIIQMNIE-ESVRKQIAKELKINQAAQCPYVV 607
Query: 65 EYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLH-VN 123
S+ + G + II+ Y + G +AD +KK + PE L Q+L L YLH
Sbjct: 608 VCYQSFYDNGV-ISIILEYMDGGSMADLLKKVKTI--PEPYLSAICKQVLKGLIYLHHER 778
Query: 124 HILHRDVKCSNIFLTKNQDIRL 145
HI+HRD+K SN+ + ++++
Sbjct: 779 HIIHRDLKPSNLLINHTGEVKI 844
Score = 56.2 bits (134), Expect(2) = 2e-23
Identities = 40/130 (30%), Positives = 66/130 (50%), Gaps = 11/130 (8%)
Frame = +3
Query: 143 IRLGDFGLAKMLTSDD-LASSIVGTPSYMCPELL--ADIPYGSKSDIWSLGCCIYEMA-- 197
+RL DFG++ ++ S A++ +GT +YM PE + + Y KSDIWSLG + E A
Sbjct: 837 LRLPDFGVSAIMESTSGQANTFIGTYNYMSPERINGSQRGYNYKSDIWSLGLILLECAMG 1016
Query: 198 --ALKPAFKAFDIQALINKINKSIVAPLPT----MYSSAFRGLVKSMLRKNPELRPPASE 251
P ++ +++ I + P P+ +SS F + + L+K+P R A E
Sbjct: 1017RFPYTPPDQSERWESIFELIETIVDKPPPSAPSEQFSSEFCSFISACLQKDPGSRLSAQE 1196
Query: 252 LLNHPHLQPY 261
L+ P + Y
Sbjct: 1197LMELPFISMY 1226
>TC78104 homologue to GP|19568098|gb|AAL89456.1 osmotic stress-activated
protein kinase {Nicotiana tabacum}, partial (95%)
Length = 1434
Score = 106 bits (265), Expect = 2e-23
Identities = 83/264 (31%), Positives = 125/264 (46%), Gaps = 8/264 (3%)
Frame = +3
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
ME+YEV++ IG G+FG A L+RHK K+L +K I + D + +E+ +R+
Sbjct: 153 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKID---ENVAREIINHRSLRH 323
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P I+ +K+ V + I++ Y G+L D I A F E++ + QL+ + Y
Sbjct: 324 PNIIRFKEV-VLTPTHLGIVMEYAAGGELFDRICSAG--RFSEDEARYFFQQLISGVSYC 494
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLG--DFGLAKMLTSDDLASSIVGTPSYMCPELLADI 178
H I HRD+K N L + RL DFG +K S VGTP+Y+ PE+L+
Sbjct: 495 HSMQICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRR 674
Query: 179 PY-GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVA-----PLPTMYSSAFR 232
Y G +D+WS G +Y M F+ D K I+A P S R
Sbjct: 675 EYDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECR 854
Query: 233 GLVKSMLRKNPELRPPASELLNHP 256
L+ + +P R E+ +HP
Sbjct: 855 HLLSRIFVASPARRITIKEIKSHP 926
>TC77179 similar to GP|2665890|gb|AAB88537.1| calcium-dependent protein
kinase {Fragaria x ananassa}, partial (92%)
Length = 2243
Score = 106 bits (265), Expect = 2e-23
Identities = 89/313 (28%), Positives = 155/313 (49%), Gaps = 14/313 (4%)
Frame = +1
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLK-----KIRLARQSDRTRRSAHQEMELI 55
++QYE+ ++G+G FG L + + + K K+R A + RR E+E++
Sbjct: 385 LQQYELGRELGRGEFGITYLCKDRETGEELACKSISKDKLRTAIDIEDVRR----EVEIM 552
Query: 56 SKV-RNPFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLL 114
+ ++P IV KD++ E V +++ CE G+L D I A G ++ E + ++
Sbjct: 553 RHLPKHPNIVTLKDTY-EDDDNVHLVMELCEGGELFDRIV-AKG-HYTERAAATVVKTIV 723
Query: 115 MALDYLHVNHILHRDVKCSNIFLTKNQD---IRLGDFGLAKMLTSDDLASSIVGTPSYMC 171
+ H + ++HRD+K N ++ ++ DFGL+ D + IVG+P YM
Sbjct: 724 QVVQMCHEHGVMHRDLKPENFLFANKKETSPLKAIDFGLSITFKPGDKFNEIVGSPYYMA 903
Query: 172 PELLADIPYGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIV----APLPTMY 227
PE+L YG + DIWS G +Y + P F A Q + I +S++ P P +
Sbjct: 904 PEVLKR-NYGPEIDIWSAGVILYILLCGIPPFWAETEQGIAQAIIRSVIDFKKEPWPKVS 1080
Query: 228 SSAFRGLVKSMLRKNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYM 287
+A + L+K ML +P+ R A E+L+HP LQ ++ L R+ Q+ N +
Sbjct: 1081DNA-KDLIKKMLDPDPKRRLTAQEVLDHPWLQNAKTAPNVSLGETVRARL-MQFSVMNKL 1254
Query: 288 RRTRF-VVPDSVS 299
++T ++ D +S
Sbjct: 1255KKTALRIIADHLS 1293
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.133 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,117,499
Number of Sequences: 36976
Number of extensions: 308352
Number of successful extensions: 2693
Number of sequences better than 10.0: 509
Number of HSP's better than 10.0 without gapping: 2417
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2470
length of query: 601
length of database: 9,014,727
effective HSP length: 102
effective length of query: 499
effective length of database: 5,243,175
effective search space: 2616344325
effective search space used: 2616344325
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0322.12


