

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0321.4
(1171 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
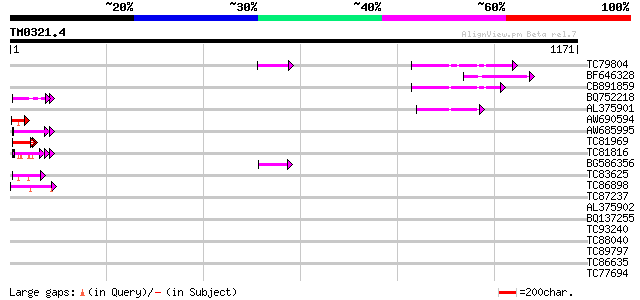
Score E
Sequences producing significant alignments: (bits) Value
TC79804 weakly similar to GP|6175165|gb|AAF04891.1| Mutator-like... 97 4e-20
BF646328 similar to PIR|F84533|F845 Mutator-like transposase [im... 86 6e-17
CB891859 similar to GP|6175165|gb| Mutator-like transposase {Ara... 83 5e-16
BQ752218 similar to GP|19170914|emb hypothetical protein {Enceph... 58 2e-08
AL375901 similar to GP|9759134|dbj mutator-like transposase-like... 53 8e-07
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 52 1e-06
AW685995 similar to PIR|I51618|I516 nucleolar phosphoprotein - A... 52 1e-06
TC81969 similar to GP|6523547|emb|CAB62280.1 hydroxyproline-rich... 52 1e-06
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 50 5e-06
BG586356 similar to GP|9759134|dbj mutator-like transposase-like... 49 1e-05
TC83625 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 45 2e-04
TC86898 similar to GP|18376007|emb|CAB91741. probable ATP citrat... 43 8e-04
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 42 0.001
AL375902 similar to GP|9759134|dbj mutator-like transposase-like... 42 0.001
BQ137255 homologue to PIR|B34768|B347 ORF5 protein - Orf virus (... 42 0.002
TC93240 weakly similar to GP|14335170|gb|AAK59865.1 AT5g51070/K3... 42 0.002
TC88040 homologue to SP|Q41266|AOX2_SOYBN Alternative oxidase 2 ... 41 0.002
TC89797 GP|13775543|gb|AAK39351.1 Hypothetical protein Y73B3A.17... 40 0.004
TC86635 weakly similar to GP|9758600|dbj|BAB09233.1 gene_id:MDF2... 40 0.007
TC77694 homologue to PIR|T09704|T09704 probable arginine/serine-... 40 0.007
>TC79804 weakly similar to GP|6175165|gb|AAF04891.1| Mutator-like transposase
{Arabidopsis thaliana}, partial (57%)
Length = 2129
Score = 97.1 bits (240), Expect = 4e-20
Identities = 61/220 (27%), Positives = 110/220 (49%)
Frame = +1
Query: 830 QGLVPTLQELIPDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMK 889
+G+V ++ P H C++H+ N K++ S L LW AA A+T+ ++++M ++
Sbjct: 976 KGIVDAIRRKFPRSSHAFCMRHLSENIGKEFKNSRLIHLLWSAAYATTINAFREKMAEIE 1155
Query: 890 AFSEEAYNDMMRLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLK 949
S A + QWA Y TR +N+ E FN +LE ++ PII ++E ++
Sbjct: 1156 EVSPNASMWLQHFHPSQWA-LVYFEGTRYGHLSSNI-EEFNKWILEAQELPIIQVIERIQ 1329
Query: 950 FYINNRIVKQRQLMLRWKTSIICPLIQQRLEFSKEAADKCVAVWNGSDNFRHFQVKRDSD 1009
+ +R W S++ P ++R+ EA ++ + F+V
Sbjct: 1330 SKLKTEFDDRRLKSSSW-CSVLTPSSERRM---VEAINRASTYQVLKSDEVEFEVISADR 1497
Query: 1010 SYVVDLEERTCACRRWQLTGIPCSHSIACMWFARRNPENY 1049
S +V++ +C+CR WQL GIPCSH++A + +R++ Y
Sbjct: 1498 SDIVNIGSHSCSCRDWQLYGIPCSHAVAALISSRKDVYAY 1617
Score = 52.4 bits (124), Expect = 1e-06
Identities = 27/76 (35%), Positives = 41/76 (53%), Gaps = 1/76 (1%)
Frame = +1
Query: 512 NFVLGQVFDSKDIFIHAVKEFALQHKKDLKIVKNDKKRVVVNCV-KGCPFNLRISKTNSR 570
NFV+GQ F F +A+KE A+ +L+I+K+D R C +GCP+ +R K +
Sbjct: 172 NFVVGQEFPDVKAFRNAIKEAAIAQHFELRIIKSDLIRYFAKCASEGCPWRIRAVKLPNA 351
Query: 571 HYFLLVTYEAAHNCCR 586
F + + E H C R
Sbjct: 352 STFTIRSLEGTHTCGR 399
>BF646328 similar to PIR|F84533|F845 Mutator-like transposase [imported] -
Arabidopsis thaliana, partial (3%)
Length = 636
Score = 86.3 bits (212), Expect = 6e-17
Identities = 51/156 (32%), Positives = 81/156 (51%), Gaps = 8/156 (5%)
Frame = -1
Query: 937 RDKPIISLLEGLKFYIN--NRIVKQRQLMLRWKTSIICPLIQQRLEFSKEAADKC---VA 991
R+KP+I LLEG + N + K ++W + + F E C A
Sbjct: 630 REKPVIXLLEGYQALYNCEDCQAKADA*KVQW*D------MSKDSSFD*EKQKSCRGWKA 469
Query: 992 VWNGSDNFRHFQVKRDSDSYVVDLEERTCACRRWQLTGIPCSHSIACMWFARRNPENYVD 1051
W+G +F V +++ Y+V+L +RTCACR+W LTGIPC+H I C+W EN+V
Sbjct: 468 TWHGDMEMNNFNVSNETNKYIVNLAQRTCACRKWNLTGIPCAHVIPCIWHNGLADENFVS 289
Query: 1052 PSYR*I---TFVVLN*LCFIVVLFVIVVVQFVILGN 1084
YR* T +L+ + F ++++ Q+++ GN
Sbjct: 288 SYYR*CSVSTIFILSKMYFYA--YMLIWYQYLVAGN 187
>CB891859 similar to GP|6175165|gb| Mutator-like transposase {Arabidopsis
thaliana}, partial (27%)
Length = 625
Score = 83.2 bits (204), Expect = 5e-16
Identities = 53/194 (27%), Positives = 99/194 (50%)
Frame = +1
Query: 830 QGLVPTLQELIPDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMK 889
QG+V ++ P H C++H+ +FRK++ + L LW+AA T+ E++ ++ ++
Sbjct: 58 QGIVDGVEANFPTAFHGFCMRHLSDSFRKEFNNTMLVNLLWEAANCLTIIEFEGKVMEIE 237
Query: 890 AFSEEAYNDMMRLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLK 949
S++A + R+P + WA +AY R N+ EA N +LE PII ++E ++
Sbjct: 238 EISQDAAYWIRRVPPRLWA-TAYFEGHRFGHLTANIVEALNSWILEASGLPIIQMMECIR 414
Query: 950 FYINNRIVKQRQLMLRWKTSIICPLIQQRLEFSKEAADKCVAVWNGSDNFRHFQVKRDSD 1009
+ ++R+ ++W TSI+ P ++ + EA ++ N F+V
Sbjct: 415 RQLMTWFNERRETSMQW-TSILVPSAERSV---AEALERARTYQVLRANEAEFEVISHEG 582
Query: 1010 SYVVDLEERTCACR 1023
+ +VD+ R C CR
Sbjct: 583 TNIVDIRNRCCLCR 624
>BQ752218 similar to GP|19170914|emb hypothetical protein {Encephalitozoon
cuniculi}, partial (5%)
Length = 637
Score = 57.8 bits (138), Expect = 2e-08
Identities = 38/88 (43%), Positives = 52/88 (58%), Gaps = 2/88 (2%)
Frame = -3
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRR 66
R R RRRRR RRR +RRRRR + +RR RR RQ R+P RRR + + R +R
Sbjct: 629 RGRKRRRRRLRRRPKRRRRRPGKKLQRRPRRPGRQPIRRPRRRRRRPKRRPRKRRGRLKR 450
Query: 67 RNHLRRRQP--RGKRLRRTWTVTRKKQV 92
R+ RRR+P R +R+RR+ + Q+
Sbjct: 449 RSRRRRRRPKRRPRRIRRSRIAKTRNQL 366
Score = 50.4 bits (119), Expect = 4e-06
Identities = 38/80 (47%), Positives = 48/80 (59%), Gaps = 2/80 (2%)
Frame = -3
Query: 6 GRRR*RRRRRRRRRRRRRRRRRR*R-RRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGP 64
GR+ RR RRRRRR +RR R+RR R +RR RRRRRR P+RR +R S +
Sbjct: 533 GRQPIRRPRRRRRRPKRRPRKRRGRLKRRSRRRRRR------PKRRPRRIRRSRI----A 384
Query: 65 RRRNHL-RRRQPRGKRLRRT 83
+ RN L R PR ++ RRT
Sbjct: 383 KTRNQLCHGRIPRTRKSRRT 324
Score = 39.7 bits (91), Expect = 0.007
Identities = 24/52 (46%), Positives = 31/52 (59%)
Frame = -3
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSST 58
RR +RR RRRRRR +RR RR RR R + R + + PR R RRT ++
Sbjct: 470 RRGRLKRRSRRRRRRPKRRPRRIRRSRIAKTRNQLCHGRIPRTRKSRRTGAS 315
>AL375901 similar to GP|9759134|dbj mutator-like transposase-like protein
{Arabidopsis thaliana}, partial (27%)
Length = 493
Score = 52.8 bits (125), Expect = 8e-07
Identities = 32/139 (23%), Positives = 64/139 (46%)
Frame = +3
Query: 841 PDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMKAFSEEAYNDMM 900
P H C++++ NFR + ++L W A A T E++ ++ M S++ +
Sbjct: 12 PSASHGFCLRYVSENFRDTFKNTKLVNIFWNAVYALTAAEFESKITEMIEVSQDVISWFQ 191
Query: 901 RLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLKFYINNRIVKQR 960
P WA AY R + E LE + P++ ++E ++ + + +R
Sbjct: 192 HFPPFLWA-VAYFDGVRYGHFTLGVTELLYNWALECHELPVVQMMEYIRQQMTSWFNDRR 368
Query: 961 QLMLRWKTSIICPLIQQRL 979
++ + W TSI+ P ++R+
Sbjct: 369 EVGMEW-TSILVPSAEKRI 422
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 52.4 bits (124), Expect = 1e-06
Identities = 34/51 (66%), Positives = 34/51 (66%), Gaps = 15/51 (29%)
Frame = +2
Query: 5 GGRRR*RRRRRR------------RRRRRRRRRRRR---*RRRRRRRRRRR 40
GGRRR* RRRRR RRRRRRRRRRR *RRRRRRRRRRR
Sbjct: 59 GGRRR*SRRRRR*T*RGRRR*T**GRRRRRRRRRRR*T**RRRRRRRRRRR 211
Score = 40.4 bits (93), Expect = 0.004
Identities = 34/60 (56%), Positives = 36/60 (59%), Gaps = 6/60 (10%)
Frame = +2
Query: 1 EENEGGRRR*RRRRRRRRRRRR--RRRRRR*----RRRRRRRRRRRQR*RKPPRRR*QRR 54
E E R * RRR RRRRR R RRR* RRRRRRRRRRR * + RRR +RR
Sbjct: 29 ETTESRRT**GGRRR*SRRRRR*T*RGRRR*T**GRRRRRRRRRRR*T**RRRRRRRRRR 208
Score = 30.4 bits (67), Expect = 4.2
Identities = 28/67 (41%), Positives = 31/67 (45%), Gaps = 14/67 (20%)
Frame = +2
Query: 2 ENEGGRRR*RRRRRRRRRRRRRRRRRR*RR--------------RRRRRRRRRQR*RKPP 47
EN+ GR RR RRR R RR RRRRRRRRR+R*
Sbjct: 2 ENDYGRIELETTESRRT**GGRRR*SRRRRR*T*RGRRR*T**GRRRRRRRRRRR*T**R 181
Query: 48 RRR*QRR 54
RRR +RR
Sbjct: 182RRRRRRR 202
>AW685995 similar to PIR|I51618|I516 nucleolar phosphoprotein - African
clawed frog, partial (5%)
Length = 516
Score = 52.0 bits (123), Expect = 1e-06
Identities = 38/85 (44%), Positives = 44/85 (51%), Gaps = 1/85 (1%)
Frame = +1
Query: 8 RR*RRRRRRR-RRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRR 66
RR*R R RR RR RR R RR R RRRRR RR+R R PP R +R + PR
Sbjct: 43 RR*RLRSLRRWRRSRRPRERRLPRPRRRRRSSRRRRRRSPPPWRSRRTAPAPRSLPAPRM 222
Query: 67 RNHLRRRQPRGKRLRRTWTVTRKKQ 91
R LRR P + L R R+ +
Sbjct: 223 RPPLRRLPPPRRPLPRRSPAPRRSR 297
Score = 43.5 bits (101), Expect = 5e-04
Identities = 35/77 (45%), Positives = 36/77 (46%), Gaps = 2/77 (2%)
Frame = +1
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRR--RQR*RKPPRRR*QRRTSSTVLRRGP 64
RR R RR R RR R RRRRR RRRRRR R R P R LRR P
Sbjct: 67 RRWRRSRRPRERRLPRPRRRRRSSRRRRRRSPPPWRSRRTAPAPRSLPAPRMRPPLRRLP 246
Query: 65 RRRNHLRRRQPRGKRLR 81
R L RR P +R R
Sbjct: 247 PPRRPLPRRSPAPRRSR 297
Score = 40.0 bits (92), Expect = 0.005
Identities = 42/99 (42%), Positives = 45/99 (45%), Gaps = 23/99 (23%)
Frame = +1
Query: 8 RR*RRRR----RRRRRRRRRRRRR-----R*RR-----------RRRRRRRRRQR*RKP- 46
RR R RR RRRRR RRRRRR R RR R R RR R+P
Sbjct: 85 RRPRERRLPRPRRRRRSSRRRRRRSPPPWRSRRTAPAPRSLPAPRMRPPLRRLPPPRRPL 264
Query: 47 PRRR*QRRTSSTVLRRGPRRRNHLRRRQP--RGKRLRRT 83
PRR R S L PRRR RR P R + LRR+
Sbjct: 265 PRRSPAPRRSRARLSPSPRRRRSPPRRPPPRRSRALRRS 381
Score = 37.7 bits (86), Expect = 0.026
Identities = 27/68 (39%), Positives = 29/68 (41%)
Frame = +1
Query: 14 RRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRR 73
RR RR RR R R RRRR R PPRR R S RR P RR ++
Sbjct: 253 RRPLPRRSPAPRRSRARLSPSPRRRRSPPRRPPPRRSRALRRSRARPRRSPMRRRRMKPA 432
Query: 74 QPRGKRLR 81
KR R
Sbjct: 433 AAENKRKR 456
Score = 31.2 bits (69), Expect = 2.4
Identities = 28/67 (41%), Positives = 30/67 (43%), Gaps = 3/67 (4%)
Frame = +1
Query: 20 RRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRGK- 78
R R RR R R RR RR RR R R+ PR R RRR RRR+ R
Sbjct: 28 RARPARR*RLRSLRRWRRSRRPRERRLPRPR--------------RRRRSSRRRRRRSPP 165
Query: 79 --RLRRT 83
R RRT
Sbjct: 166PWRSRRT 186
Score = 29.6 bits (65), Expect = 7.1
Identities = 23/62 (37%), Positives = 28/62 (45%), Gaps = 5/62 (8%)
Frame = +1
Query: 12 RRRRRR-----RRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRR 66
RR R R RRRR RR RR R RR R + R P RRR + ++ R+
Sbjct: 286 RRSRARLSPSPRRRRSPPRRPPPRRSRALRRSRARPRRSPMRRRRMKPAAAENKRKRDEE 465
Query: 67 RN 68
N
Sbjct: 466 EN 471
>TC81969 similar to GP|6523547|emb|CAB62280.1 hydroxyproline-rich
glycoprotein DZ-HRGP {Volvox carteri f. nagariensis},
partial (11%)
Length = 556
Score = 52.0 bits (123), Expect = 1e-06
Identities = 31/48 (64%), Positives = 36/48 (74%)
Frame = +1
Query: 6 GRRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QR 53
GR R RR +RRRR+R+ RRRR +RRRRRRRRRR+R R RRR QR
Sbjct: 238 GRTRRRRFKRRRRKRQIWRRRRMGKRRRRRRRRRRRRRR---RRRVQR 372
Score = 50.1 bits (118), Expect = 5e-06
Identities = 32/54 (59%), Positives = 38/54 (70%), Gaps = 3/54 (5%)
Frame = +1
Query: 7 RRR*RRRRRRRRRRRR---RRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSS 57
R R RRRR +RRRR+R RRRR RRRRRRRRRRR+R R+ +R *+ R S
Sbjct: 235 RGRTRRRRFKRRRRKRQIWRRRRMGKRRRRRRRRRRRRRRRRRVQRI*RDRRIS 396
Score = 42.4 bits (98), Expect = 0.001
Identities = 22/28 (78%), Positives = 24/28 (85%)
Frame = +1
Query: 6 GRRR*RRRRRRRRRRRRRRRRRR*RRRR 33
G+RR RRRRRRRRRRRRRR +R *R RR
Sbjct: 307 GKRRRRRRRRRRRRRRRRRVQRI*RDRR 390
Score = 40.8 bits (94), Expect = 0.003
Identities = 31/75 (41%), Positives = 39/75 (51%)
Frame = +1
Query: 15 RRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQ 74
R RR R R RRRR RRRR+R+ RR+R K RRR RRR RRR+
Sbjct: 220 RTRRIRGRTRRRRFKRRRRKRQIWRRRRMGKRRRRR--------------RRRRRRRRRR 357
Query: 75 PRGKRLRRTWTVTRK 89
R +R+ R ++ K
Sbjct: 358 RRVQRI*RDRRISLK 402
Score = 36.6 bits (83), Expect = 0.058
Identities = 34/88 (38%), Positives = 44/88 (49%), Gaps = 4/88 (4%)
Frame = +1
Query: 9 R*RRRRRRRRRRRRRRRRRR*RRR----RRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGP 64
R* R RRR R R R *+ R R RR R R R R+ RRR R+ RR
Sbjct: 136 R*GWRSRRRWTRWRLGRGNA*QCRLGNFRTRRIRGRTRRRRFKRRR--RKRQIWRRRRMG 309
Query: 65 RRRNHLRRRQPRGKRLRRTWTVTRKKQV 92
+RR RRR+ R +R RR + R +++
Sbjct: 310 KRRRRRRRRRRRRRRRRRVQRI*RDRRI 393
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 50.1 bits (118), Expect = 5e-06
Identities = 29/71 (40%), Positives = 41/71 (56%)
Frame = +1
Query: 11 RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHL 70
RR RRRR+R+RRRR ++ R +RR R+ R RK R + + V +R R R
Sbjct: 319 RRIRRRRKRKRRRREKKTKTRMVKRRNLRKTRRRKRTRMKTMMKVRMEVRKRRIRIRKRK 498
Query: 71 RRRQPRGKRLR 81
RRR+ R KR++
Sbjct: 499 RRRRMRKKRVK 531
Score = 48.5 bits (114), Expect = 1e-05
Identities = 35/101 (34%), Positives = 57/101 (55%), Gaps = 15/101 (14%)
Frame = +1
Query: 7 RRR*RRRRRRR--------RRRRRRRRRRR*RRRRRRR-------RRRRQR*RKPPRRR* 51
R+R RR RRRR R RR +RRR++ RR+R R +RR +R RK RRR
Sbjct: 181 RKRKRRIRRRRTKLM*MRVRIRRTKRRRKKKRRKRMLRVKKKMVMKRRIRRRRKRKRRRR 360
Query: 52 QRRTSSTVLRRGPRRRNHLRRRQPRGKRLRRTWTVTRKKQV 92
+++T + +++R R+ RR++ R K + + RK+++
Sbjct: 361 EKKTKTRMVKRRNLRKTR-RRKRTRMKTMMKVRMEVRKRRI 480
Score = 47.0 bits (110), Expect = 4e-05
Identities = 32/92 (34%), Positives = 51/92 (54%), Gaps = 17/92 (18%)
Frame = +1
Query: 8 RR*RRRRRRRRRRR-----------RRRRRRR*RRRRRRRRR------RRQR*RKPPRRR 50
RR +RRR+++RR+R RR RRRR R+RRRR ++ +R+ RK RR+
Sbjct: 244 RRTKRRRKKKRRKRMLRVKKKMVMKRRIRRRRKRKRRRREKKTKTRMVKRRNLRKTRRRK 423
Query: 51 *QRRTSSTVLRRGPRRRNHLRRRQPRGKRLRR 82
R + +R R+R R++ R +R+R+
Sbjct: 424 RTRMKTMMKVRMEVRKRRIRIRKRKRRRRMRK 519
Score = 42.7 bits (99), Expect = 8e-04
Identities = 31/83 (37%), Positives = 45/83 (53%), Gaps = 17/83 (20%)
Frame = +1
Query: 7 RRR*RRRRRRRRRRRR---------RRRRRR*RRRRRRRRRRRQR*R--------KPPRR 49
RR R+ RRR+R R + R+RR R R+R+RRRR R++R + K PR+
Sbjct: 391 RRNLRKTRRRKRTRMKTMMKVRMEVRKRRIRIRKRKRRRRMRKKRVKYL*GILI*KKPRK 570
Query: 50 R*QRRTSSTVLRRGPRRRNHLRR 72
+ +RR R+ RRR +L R
Sbjct: 571 KVKRRRRKKKTRK--RRRRNLER 633
Score = 41.2 bits (95), Expect = 0.002
Identities = 28/86 (32%), Positives = 45/86 (51%), Gaps = 5/86 (5%)
Frame = +1
Query: 2 ENEGGRRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTS----- 56
E + R +RR R+ RRR+R R + + R R+RR R RK RRR R+
Sbjct: 361 EKKTKTRMVKRRNLRKTRRRKRTRMKTMMKVRMEVRKRRIRIRKRKRRRRMRKKRVKYL* 540
Query: 57 STVLRRGPRRRNHLRRRQPRGKRLRR 82
++ + PR++ RRR+ + ++ RR
Sbjct: 541 GILI*KKPRKKVKRRRRKKKTRKRRR 618
Score = 30.8 bits (68), Expect = 3.2
Identities = 20/59 (33%), Positives = 29/59 (48%), Gaps = 17/59 (28%)
Frame = +1
Query: 7 RRR*RRRRRRRRRRR-----------------RRRRRRR*RRRRRRRRRRRQR*RKPPR 48
R R R+RRRR R++R +RRRR++ R+RRRR R + P +
Sbjct: 481 RIRKRKRRRRMRKKRVKYL*GILI*KKPRKKVKRRRRKKKTRKRRRRNLERIKTNDPSK 657
>BG586356 similar to GP|9759134|dbj mutator-like transposase-like protein
{Arabidopsis thaliana}, partial (20%)
Length = 696
Score = 48.5 bits (114), Expect = 1e-05
Identities = 24/72 (33%), Positives = 39/72 (53%), Gaps = 1/72 (1%)
Frame = +3
Query: 514 VLGQVFDSKDIFIHAVKEFALQHKKDLKIVKNDKKRVVVNCVK-GCPFNLRISKTNSRHY 572
V+GQ F + +K+ A+ DL+IVK+D+ R + C K GCP+ + ++K
Sbjct: 330 VIGQEFPDVETCRRTLKDIAIAMHFDLRIVKSDRSRFIAKCSKEGCPWRVHVAKCPGVPT 509
Query: 573 FLLVTYEAAHNC 584
F + T +A H C
Sbjct: 510 FTIRTLQADHTC 545
>TC83625 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (9%)
Length = 908
Score = 45.1 bits (105), Expect = 2e-04
Identities = 31/82 (37%), Positives = 47/82 (56%), Gaps = 15/82 (18%)
Frame = +3
Query: 7 RRR*RRRRRRR--------RRRRRRRRRRR*RRRRRRR-------RRRRQR*RKPPRRR* 51
R+R RR RRRR R RR +RRR++ RR+R R +RR +R RK RRR
Sbjct: 573 RKRKRRIRRRRTKLM*MRVRIRRTKRRRKKKRRKRMLRVKKKMVMKRRIRRRRKRKRRRR 752
Query: 52 QRRTSSTVLRRGPRRRNHLRRR 73
+++T + +++R R+ R+R
Sbjct: 753 EKKTKTRMVKRRNLRKTRRRKR 818
Score = 40.4 bits (93), Expect = 0.004
Identities = 27/80 (33%), Positives = 41/80 (50%), Gaps = 8/80 (10%)
Frame = +3
Query: 7 RRR*RRRRRRRRRRRRRRRRR--------R*RRRRRRRRRRRQR*RKPPRRR*QRRTSST 58
R R RR +RRR+++RR+R R R RRRR+R+RRR+ + R +R T
Sbjct: 624 RVRIRRTKRRRKKKRRKRMLRVKKKMVMKRRIRRRRKRKRRRREKKTKTRMVKRRNLRKT 803
Query: 59 VLRRGPRRRNHLRRRQPRGK 78
R+ R + + + GK
Sbjct: 804 RRRKRTRMKTMMEDEKEEGK 863
>TC86898 similar to GP|18376007|emb|CAB91741. probable ATP citrate lyase
subunit 2 {Neurospora crassa}, complete
Length = 2389
Score = 42.7 bits (99), Expect = 8e-04
Identities = 42/122 (34%), Positives = 52/122 (42%), Gaps = 26/122 (21%)
Frame = -3
Query: 2 ENEGGRRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRR-----------------QR*R 44
E++ R R RRR RRRR RRRRRRR RRR R R +R +
Sbjct: 1670 EHQSTHRSWRARRRSRRRRWRRRRRRRHHHRRRESRYGRWG*GQ*A*GRHRSWHQGRRCK 1491
Query: 45 KPP---RRR*QRRTSSTVLRRG-PRRRNHLRRRQPRGKRLRRTW-----TVTRKKQVVWS 95
PP R +R T S R P RR +R RG+R W T +R+ W
Sbjct: 1490 PPPWSAHDRKERGTPSAGPHRC*PFRRWKRQRWSGRGQRESGQWPT*HRTQSRRSGRAWQ 1311
Query: 96 QN 97
Q+
Sbjct: 1310 QD 1305
Score = 36.6 bits (83), Expect = 0.058
Identities = 20/37 (54%), Positives = 22/37 (59%)
Frame = -3
Query: 6 GRRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR 42
G R + R R RRR RRRR RRRRRRR R+R
Sbjct: 1682 GTRWEHQSTHRSWRARRRSRRRRWRRRRRRRHHHRRR 1572
Score = 32.7 bits (73), Expect = 0.84
Identities = 20/43 (46%), Positives = 22/43 (50%)
Frame = -3
Query: 6 GRRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPR 48
GR R + R R RRR R RR RRRRRRR R+ R
Sbjct: 1691 GRSGTRWEHQSTHRSWRARRRSRRRRWRRRRRRRHHHRRRESR 1563
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 42.0 bits (97), Expect = 0.001
Identities = 35/137 (25%), Positives = 65/137 (46%), Gaps = 8/137 (5%)
Frame = +1
Query: 303 EEVVSVEKEVEKDIGSVVLECERQVEKEAEKEIGSVVLE---CERQVEKEAEKENQVVGE 359
EE +E E K+ G E + +E E KE G E E + K+ EK Q E
Sbjct: 1 EEKSHLENEESKETGESSEE-KSHLENEESKETGESSEEKSHLENEENKDEEKSKQENEE 177
Query: 360 QAGKEKVVVEEAEKENQVVGEQAGKEKVVVEQAEKENQ---GVGGEED--DESYHPNSDD 414
EK+ E E +++ +Q +E E++++EN+ GGE++ + + + +
Sbjct: 178 IKDGEKIQQENEENKDEEKSQQENEENKDEEKSQQENELKKNEGGEKETGEITEEKSKQE 357
Query: 415 SDDSSVVSLDDSDYEEN 431
++++S + D + EE+
Sbjct: 358 NEETSETNSKDKENEES 408
Score = 38.1 bits (87), Expect = 0.020
Identities = 36/139 (25%), Positives = 54/139 (37%), Gaps = 11/139 (7%)
Frame = +1
Query: 293 KDVEKDKVIQEEVVSVEK---EVEKDIGSVVLECERQVEKEAEKEIGSVVLECERQVEKE 349
KD EK K EE+ EK E E++ + E + K+ EK L+ EKE
Sbjct: 142 KDEEKSKQENEEIKDGEKIQQENEENKDEEKSQQENEENKDEEKSQQENELKKNEGGEKE 321
Query: 350 AEKENQVVGEQAGKEKVVVEEAEKENQVVGEQAGKEKVVV--------EQAEKENQGVGG 401
+ + +Q +E +KEN+ + K V +Q +E G G
Sbjct: 322 TGEITEEKSKQENEETSETNSKDKENEESNQNGSDAKEQVGENHEQDSKQGTEETNGTEG 501
Query: 402 EEDDESYHPNSDDSDDSSV 420
E +E D S D+ V
Sbjct: 502 GEKEEHDKIKEDTSSDNQV 558
>AL375902 similar to GP|9759134|dbj mutator-like transposase-like protein
{Arabidopsis thaliana}, partial (20%)
Length = 542
Score = 42.0 bits (97), Expect = 0.001
Identities = 15/37 (40%), Positives = 22/37 (58%)
Frame = +2
Query: 1018 RTCACRRWQLTGIPCSHSIACMWFARRNPENYVDPSY 1054
R C+CRRWQL G PC+H+ A + N + +P +
Sbjct: 8 RECSCRRWQLYGYPCAHAAAALISCGHNAHMFAEPCF 118
>BQ137255 homologue to PIR|B34768|B347 ORF5 protein - Orf virus (strain NZ2),
partial (10%)
Length = 1161
Score = 41.6 bits (96), Expect = 0.002
Identities = 28/65 (43%), Positives = 32/65 (49%)
Frame = +3
Query: 4 EGGRRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRG 63
E GRR R+RRRRR R R R+R RR R R R R PRR +R S RR
Sbjct: 615 EDGRRASDDAARQRRRRRAPRAREAHRQRARRARAARDRRRPAPRR---KRARSRAARRA 785
Query: 64 PRRRN 68
R +
Sbjct: 786 RARHS 800
Score = 37.4 bits (85), Expect = 0.034
Identities = 35/106 (33%), Positives = 44/106 (41%), Gaps = 17/106 (16%)
Frame = +3
Query: 3 NEGGRRR*RRRRRRRRRRRRRRRRRR*RRRRRRRR--RRRQR*RKPPRRR*QRRTS---- 56
++ R+ RRRR R R R+R RR R R RRR RR+R R RR + R S
Sbjct: 633 SDDAARQRRRRRAPRAREAHRQRARRARAARDRRRPAPRRKRARSRAARRARARHSIQED 812
Query: 57 -----------STVLRRGPRRRNHLRRRQPRGKRLRRTWTVTRKKQ 91
S R RR RRR R R T R+++
Sbjct: 813 ADDARDDEIDESADTRAARARRRDSRRRASRATAARARQTKERRER 950
Score = 33.5 bits (75), Expect = 0.49
Identities = 23/55 (41%), Positives = 25/55 (44%)
Frame = +2
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLR 61
R R R R R R R R R R RRR R RR+R PP R R S+ R
Sbjct: 539 RPRARDARATRARNDERARGRVARSRRRTTRERRRRPTAPPTPRAARARSAQTAR 703
Score = 31.6 bits (70), Expect = 1.9
Identities = 32/87 (36%), Positives = 35/87 (39%), Gaps = 7/87 (8%)
Frame = +2
Query: 6 GRRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLR---- 61
GRR R RRR RR+RR RR R R R R R R R T T R
Sbjct: 806 GRRGRRARRRDRRKRRHARRARTPARFASARVARDGSARATDERA-TRTTGDTRTRTRRD 982
Query: 62 ---RGPRRRNHLRRRQPRGKRLRRTWT 85
RG R H R R +R R+ T
Sbjct: 983 ASARG--RSRHARSRDNDARRRERSTT 1057
Score = 31.2 bits (69), Expect = 2.4
Identities = 30/81 (37%), Positives = 35/81 (43%), Gaps = 5/81 (6%)
Frame = -3
Query: 8 RR*RRRRRRRRRRRRRRR-----RRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRR 62
RR RRR R R RR R R R RRRRR R R+P R +SS L
Sbjct: 751 RRGAGRRRSRAARARRARCLCASRARGARRRRRCRAASSLARRPSSA--PRYSSSRALVV 578
Query: 63 GPRRRNHLRRRQPRGKRLRRT 83
RR + + R R RR+
Sbjct: 577 SCARRPRIACARTRDARARRS 515
>TC93240 weakly similar to GP|14335170|gb|AAK59865.1 AT5g51070/K3K7_27
{Arabidopsis thaliana}, partial (8%)
Length = 700
Score = 41.6 bits (96), Expect = 0.002
Identities = 22/32 (68%), Positives = 24/32 (74%)
Frame = -2
Query: 4 EGGRRR*RRRRRRRRRRRRRRRRRR*RRRRRR 35
E G RR RRRRRRRRRRRRR R*++R RR
Sbjct: 261 E*GERRRRRRRRRRRRRRRRTVSLR*KKRERR 166
Score = 40.0 bits (92), Expect = 0.005
Identities = 20/28 (71%), Positives = 22/28 (78%)
Frame = -2
Query: 12 RRRRRRRRRRRRRRRRR*RRRRRRRRRR 39
RRRRRRRRRRRRRRRR R ++R RR
Sbjct: 249 RRRRRRRRRRRRRRRRTVSLR*KKRERR 166
Score = 39.3 bits (90), Expect = 0.009
Identities = 20/28 (71%), Positives = 22/28 (78%)
Frame = -2
Query: 13 RRRRRRRRRRRRRRRR*RRRRRRRRRRR 40
RRRRRRRRRRRRRRRR R ++R RR
Sbjct: 249 RRRRRRRRRRRRRRRRTVSLR*KKRERR 166
Score = 38.5 bits (88), Expect = 0.015
Identities = 29/78 (37%), Positives = 36/78 (45%)
Frame = -2
Query: 22 RRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRGKRLR 81
RRRRRRR RRRRRRRR R*+K RR * G R + + G
Sbjct: 249 RRRRRRRRRRRRRRRRTVSLR*KKRERRW*VYGGG------GGRWFR*VNAAEAVGSESW 88
Query: 82 RTWTVTRKKQVVWSQNVN 99
W KK++ W ++ N
Sbjct: 87 EPWVKVMKKKLAWFESGN 34
Score = 37.7 bits (86), Expect = 0.026
Identities = 19/27 (70%), Positives = 21/27 (77%)
Frame = -2
Query: 11 RRRRRRRRRRRRRRRRRR*RRRRRRRR 37
RRRRRRRRRRRRRRR R ++R RR
Sbjct: 246 RRRRRRRRRRRRRRRTVSLR*KKRERR 166
Score = 35.8 bits (81), Expect = 0.099
Identities = 18/27 (66%), Positives = 21/27 (77%)
Frame = -2
Query: 16 RRRRRRRRRRRRR*RRRRRRRRRRRQR 42
RRRRRRRRRRRRR RR R ++R+R
Sbjct: 249 RRRRRRRRRRRRRRRRTVSLR*KKRER 169
Score = 35.4 bits (80), Expect = 0.13
Identities = 18/28 (64%), Positives = 21/28 (74%)
Frame = -2
Query: 15 RRRRRRRRRRRRRR*RRRRRRRRRRRQR 42
RRRRRRRRRRRRRR R R ++R +R
Sbjct: 249 RRRRRRRRRRRRRRRRTVSLR*KKRERR 166
Score = 35.4 bits (80), Expect = 0.13
Identities = 18/28 (64%), Positives = 21/28 (74%)
Frame = -2
Query: 14 RRRRRRRRRRRRRRR*RRRRRRRRRRRQ 41
RRRRRRRRRRRRRRR R ++R R+
Sbjct: 249 RRRRRRRRRRRRRRRRTVSLR*KKRERR 166
Score = 30.8 bits (68), Expect = 3.2
Identities = 19/32 (59%), Positives = 21/32 (65%), Gaps = 4/32 (12%)
Frame = -2
Query: 1 EENEGGRRR*RRRRRRRRRR----RRRRRRRR 28
E E RRR RRRRRRRRRR R ++R RR
Sbjct: 261 E*GERRRRRRRRRRRRRRRRTVSLR*KKRERR 166
>TC88040 homologue to SP|Q41266|AOX2_SOYBN Alternative oxidase 2
mitochondrial precursor (EC 1.-.-.-). [Soybean] {Glycine
max}, partial (82%)
Length = 1678
Score = 41.2 bits (95), Expect = 0.002
Identities = 20/31 (64%), Positives = 23/31 (73%)
Frame = +3
Query: 12 RRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR 42
R+R RR RR+RRRR R RR RRRRRR +R
Sbjct: 567 RKRHRRSHRRKRRRRNRPRRTRRRRRRVPER 659
Score = 39.7 bits (91), Expect = 0.007
Identities = 21/32 (65%), Positives = 22/32 (68%)
Frame = +3
Query: 9 R*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRR 40
R R RR RR+RRRR R RR RRRRRR RR
Sbjct: 567 RKRHRRSHRRKRRRRNRPRRTRRRRRRVPERR 662
Score = 38.1 bits (87), Expect = 0.020
Identities = 20/32 (62%), Positives = 22/32 (68%)
Frame = +3
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRR 38
R+R RR RR+RRRR R RR R RRRR RR
Sbjct: 567 RKRHRRSHRRKRRRRNRPRRTRRRRRRVPERR 662
Score = 38.1 bits (87), Expect = 0.020
Identities = 19/32 (59%), Positives = 22/32 (68%)
Frame = +3
Query: 11 RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR 42
R+R RR RR+RRRR R R RRRRRR +R
Sbjct: 567 RKRHRRSHRRKRRRRNRPRRTRRRRRRVPERR 662
Score = 36.2 bits (82), Expect = 0.076
Identities = 19/32 (59%), Positives = 21/32 (65%)
Frame = +3
Query: 8 RR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRR 39
R+ RR RR+RRRR R RR RRRRR RR
Sbjct: 567 RKRHRRSHRRKRRRRNRPRRTRRRRRRVPERR 662
Score = 33.9 bits (76), Expect = 0.38
Identities = 25/57 (43%), Positives = 29/57 (50%)
Frame = +3
Query: 26 RRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRGKRLRR 82
R+R RR RR+RRRR R R+ RRR RR P RR RR G R +R
Sbjct: 567 RKRHRRSHRRKRRRRNRPRRTRRRR----------RRVPERRITWLRRVIGGFRGQR 707
Score = 33.9 bits (76), Expect = 0.38
Identities = 20/34 (58%), Positives = 21/34 (60%)
Frame = +3
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRR 40
RR RR+RRRR R RR RRRRR RR RR
Sbjct: 579 RRSHRRKRRRRNRPRRTRRRRRRVPERRITWLRR 680
>TC89797 GP|13775543|gb|AAK39351.1 Hypothetical protein Y73B3A.17
{Caenorhabditis elegans}, partial (1%)
Length = 958
Score = 40.4 bits (93), Expect = 0.004
Identities = 43/151 (28%), Positives = 79/151 (51%), Gaps = 7/151 (4%)
Frame = -2
Query: 261 ANEISLD------AVDSDDDVAIVDRLDVICGKLLQGEKDVEKDKVIQEEVVSVEKEVEK 314
AN ++D A+D +DV V I EK VE+ I+ EV SV++E K
Sbjct: 786 ANHAAMDVEDKNLALDLQNDVFFVFVKTQITELGFLFEKIVEEKNEIEYEV-SVQREKMK 610
Query: 315 DIGSVVLECERQVEKEAEKEIGSVVLECERQVEKEAEKENQVVGEQAGKEK-VVVEEAEK 373
D+G + ER VEK + L+ + +++AE+E++V E+ K++ + V+++ +
Sbjct: 609 DLGLCLESEERNVEK--------MRLDARKLFDEKAERESKV--EELEKDRDLAVKKSIE 460
Query: 374 ENQVVGEQAGKEKVVVEQAEKENQGVGGEED 404
+V+ E K +VV++ + + +GV +D
Sbjct: 459 SVKVIDELKEKIDLVVKE-KNDAEGVNSTQD 370
>TC86635 weakly similar to GP|9758600|dbj|BAB09233.1
gene_id:MDF20.10~pir||T08929~similar to unknown protein
{Arabidopsis thaliana}, partial (32%)
Length = 1963
Score = 39.7 bits (91), Expect = 0.007
Identities = 26/84 (30%), Positives = 45/84 (52%), Gaps = 8/84 (9%)
Frame = +3
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRR 66
R+R RR+ + RRR++ R ++R+RRR+R + RK +RR +RR + +L ++
Sbjct: 6 RKRVMMRRKLK*RRRKKVMTMRNLKKRQRRRKRAMKRRKLKQRRRRRRRPTRILLINQKK 185
Query: 67 RNHLR--------RRQPRGKRLRR 82
+ R + P GK +RR
Sbjct: 186KIRPRVVRVRKDQKSVPEGKLMRR 257
>TC77694 homologue to PIR|T09704|T09704 probable arginine/serine-rich
splicing factor - alfalfa, partial (95%)
Length = 1494
Score = 39.7 bits (91), Expect = 0.007
Identities = 32/79 (40%), Positives = 37/79 (46%), Gaps = 6/79 (7%)
Frame = +3
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRR------RRRQR*RKPPRRR*QRRTSSTVL 60
RRR R R R R R R R R RRR R R + R R R RR R S +V
Sbjct: 447 RRRSRSRSYDRHERDRHRGRDRDHRRRSRSRSASPGYKGRGRGRHDDERR-SRSPSRSVD 623
Query: 61 RRGPRRRNHLRRRQPRGKR 79
R P RR+ + +R P KR
Sbjct: 624 SRSPVRRSSIPKRSPSPKR 680
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.338 0.149 0.483
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,316,567
Number of Sequences: 36976
Number of extensions: 687457
Number of successful extensions: 24259
Number of sequences better than 10.0: 349
Number of HSP's better than 10.0 without gapping: 5187
Number of HSP's successfully gapped in prelim test: 357
Number of HSP's that attempted gapping in prelim test: 2926
Number of HSP's gapped (non-prelim): 13690
length of query: 1171
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1064
effective length of database: 5,058,295
effective search space: 5382025880
effective search space used: 5382025880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0321.4


