

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
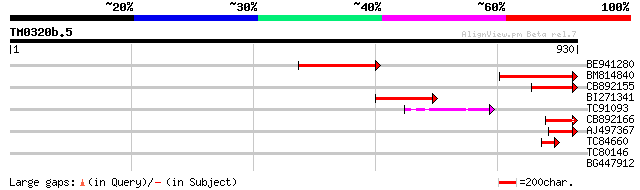
Query= TM0320b.5
(930 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BE941280 weakly similar to GP|20197614|g unknown protein {Arabid... 172 4e-43
BM814840 weakly similar to GP|15128241|db helicase-like protein ... 150 2e-36
CB892155 similar to PIR|D86481|D86 hypothetical protein AAG28292... 99 1e-20
BI271341 83 4e-16
TC91093 weakly similar to PIR|T01873|T01873 hypothetical protein... 80 5e-15
CB892166 similar to GP|20197614|gb unknown protein {Arabidopsis ... 59 9e-09
AJ497367 similar to GP|14140286|gb putative helicase {Oryza sati... 56 6e-08
TC84660 similar to PIR|D86481|D86481 hypothetical protein AAG282... 44 4e-04
TC80146 40 0.003
BG447912 31 2.5
>BE941280 weakly similar to GP|20197614|g unknown protein {Arabidopsis
thaliana}, partial (8%)
Length = 403
Score = 172 bits (437), Expect = 4e-43
Identities = 86/134 (64%), Positives = 102/134 (75%)
Frame = -1
Query: 474 LYGFGGSGKTFVWNTLSAALRSEGKIVLNVASSGIASLLLPGGRTAHSRFSIPITIHESS 533
LY +GG+ KTF+W LSAALRSEG+IVL ASSGI +LL+PGGRTAHSRF IP I E+S
Sbjct: 403 LYDYGGTEKTFIWRALSAALRSEGEIVLACASSGIDALLMPGGRTAHSRFGIPFIIDETS 224
Query: 534 TCNVRQGSHKAEMLQKASLIIWDEAPMLNKHCFEALDKTLNDIMKTQATFGHDKPFAGKV 593
C V A ++ KA LIIWDEAPM++KHCFEALD++L D++KT D PF GKV
Sbjct: 223 MCGVTPNIPLASLVIKAKLIIWDEAPMMHKHCFEALDRSLRDVLKTVDERNKDIPFGGKV 44
Query: 594 VVLGGDFRQILPVI 607
VVLGGDFRQIL V+
Sbjct: 43 VVLGGDFRQILLVM 2
>BM814840 weakly similar to GP|15128241|db helicase-like protein {Oryza
sativa (japonica cultivar-group)}, partial (6%)
Length = 733
Score = 150 bits (379), Expect = 2e-36
Identities = 76/127 (59%), Positives = 94/127 (73%)
Frame = +3
Query: 804 NGTRLIVSALTPYIIVATALSGSKTGKPVYIPRLSLTPSDTGLPFKFSRRQFPITVCFAM 863
+GTRLI+ +L +I A + G+ G+ YIPR++L PS + F R QFP+ + FAM
Sbjct: 27 HGTRLIIVSLGKNVICARVIGGTHAGEVSYIPRMNLIPSGANVSITFERCQFPLVLSFAM 206
Query: 864 TINKSQGQSLSHVGLYLPRPVFTHGQLYVALSRVKSRKRLKILIVDDKGVVSNCTRNVVY 923
TINKSQGQ+L+ VGLYLPRPVFTHGQLYVA+SRVKSR LKILI D+ G S+ T NVVY
Sbjct: 207 TINKSQGQTLTSVGLYLPRPVFTHGQLYVAVSRVKSRSGLKILITDENGSPSSSTVNVVY 386
Query: 924 EEVFQNI 930
+EVFQ I
Sbjct: 387 QEVFQKI 407
>CB892155 similar to PIR|D86481|D86 hypothetical protein AAG28292.1
[imported] - Arabidopsis thaliana, partial (4%)
Length = 572
Score = 98.6 bits (244), Expect = 1e-20
Identities = 50/74 (67%), Positives = 57/74 (76%)
Frame = -2
Query: 857 ITVCFAMTINKSQGQSLSHVGLYLPRPVFTHGQLYVALSRVKSRKRLKILIVDDKGVVSN 916
+ V FAMTINKSQGQSL H+G+YLP VF+HGQLYVALSRV SR+ LKILI +D G
Sbjct: 370 VEVYFAMTINKSQGQSLKHIGVYLPSSVFSHGQLYVALSRVTSREGLKILISNDDGEDDC 191
Query: 917 CTRNVVYEEVFQNI 930
T NVVY EVF N+
Sbjct: 190 VTSNVVYREVFHNL 149
>BI271341
Length = 468
Score = 83.2 bits (204), Expect = 4e-16
Identities = 46/103 (44%), Positives = 67/103 (64%)
Frame = +1
Query: 600 FRQILPVILKGSRSEIISSSANSSYLWKHCKVMKLTTNMRLQQAGSSSSSLEIKEFADWL 659
FR ILPVI +GSRS+II ++ NSS + HC+V++L NM LQQ G SS+ E ++F+ +
Sbjct: 1 FR*ILPVIPRGSRSDIIHATINSSCI*DHCQVVRLKKNMWLQQNGQSSNDPEFEQFSK*I 180
Query: 660 LQVGDGTIKPIDEDDSIIEIPTYLLVRESDNPLLELVNFAYLN 702
L+VGDG I ++ + I+IP LL+ D+ L +V Y N
Sbjct: 181 LKVGDGKIYEPNDSYADIDIPPELLISNYDDSLQTIVQSTYQN 309
>TC91093 weakly similar to PIR|T01873|T01873 hypothetical protein T24M8.10 -
Arabidopsis thaliana, partial (11%)
Length = 701
Score = 79.7 bits (195), Expect = 5e-15
Identities = 49/148 (33%), Positives = 81/148 (54%)
Frame = +3
Query: 648 SSLEIKEFADWLLQVGDGTIKPIDEDDSIIEIPTYLLVRESDNPLLELVNFAYLNVVANL 707
+ +E K FA+ L T K + +DS E+ T +P+ +V Y N+V+
Sbjct: 297 TKVEFKTFAEIL------TGKMSEPNDSYAEVDT-----PPGDPIDAIVQSTYPNLVSQY 443
Query: 708 ENHAYFEQRALLAPTLESVEEVNNFMMSMIPGEETKYLSYDTPCRSDEDSEIDAEWFTSE 767
N + + RA+L T E V+++N++++ +IPGEE S + RS+ + + E
Sbjct: 444 NNEQFLQSRAILTSTDEVVDQINDYVLKLIPGEERVIYSAN---RSEVNDVQAFDAIPPE 614
Query: 768 FLNDLKCSGIPNHRIVLKVGVPIMLIQN 795
FL LK S +PNH++ LKVG PIML+++
Sbjct: 615 FLQSLKTSDLPNHKLTLKVGTPIMLLRD 698
>CB892166 similar to GP|20197614|gb unknown protein {Arabidopsis thaliana},
partial (3%)
Length = 748
Score = 58.9 bits (141), Expect = 9e-09
Identities = 30/52 (57%), Positives = 39/52 (74%)
Frame = -1
Query: 879 YLPRPVFTHGQLYVALSRVKSRKRLKILIVDDKGVVSNCTRNVVYEEVFQNI 930
+ R VF+HGQLYVA+SRV SR LKIL++D+ G + T NVVY +VFQN+
Sbjct: 289 FYDREVFSHGQLYVAISRVSSRNGLKILMIDENGDCIDNTTNVVY-KVFQNV 137
>AJ497367 similar to GP|14140286|gb putative helicase {Oryza sativa (japonica
cultivar-group)}, partial (1%)
Length = 543
Score = 56.2 bits (134), Expect = 6e-08
Identities = 27/46 (58%), Positives = 36/46 (77%)
Frame = +1
Query: 885 FTHGQLYVALSRVKSRKRLKILIVDDKGVVSNCTRNVVYEEVFQNI 930
F++G+LYVA+SRV SRK LKIL+ + G N T NVVY+EVF+N+
Sbjct: 19 FSNGKLYVAVSRVTSRKGLKILLAHEDGNCMNTTSNVVYKEVFRNL 156
>TC84660 similar to PIR|D86481|D86481 hypothetical protein AAG28292.1
[imported] - Arabidopsis thaliana, partial (1%)
Length = 1009
Score = 43.5 bits (101), Expect = 4e-04
Identities = 20/30 (66%), Positives = 25/30 (82%)
Frame = +2
Query: 872 SLSHVGLYLPRPVFTHGQLYVALSRVKSRK 901
S++ G+YLP+P+F HG LYVALSRV SRK
Sbjct: 68 SMATKGMYLPQPIF*HG*LYVALSRVTSRK 157
>TC80146
Length = 476
Score = 40.4 bits (93), Expect = 0.003
Identities = 17/35 (48%), Positives = 27/35 (76%)
Frame = -3
Query: 857 ITVCFAMTINKSQGQSLSHVGLYLPRPVFTHGQLY 891
I + + TINKS+ QSLS++ +YL RP+F+H ++Y
Sbjct: 471 IQIPYFKTINKSR*QSLSYMKIYLSRPIFSHEEMY 367
>BG447912
Length = 645
Score = 30.8 bits (68), Expect = 2.5
Identities = 18/56 (32%), Positives = 26/56 (46%)
Frame = -3
Query: 738 PGEETKYLSYDTPCRSDEDSEIDAEWFTSEFLNDLKCSGIPNHRIVLKVGVPIMLI 793
PG E ++ D C ++DA SEFL DLK + PN + G+ M +
Sbjct: 439 PGREASRINLDGRC-----PDVDAAAVESEFLTDLKTNFEPNLLPLFPAGIKAMTV 287
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.344 0.151 0.484
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,023,914
Number of Sequences: 36976
Number of extensions: 384508
Number of successful extensions: 3363
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 1407
Number of HSP's successfully gapped in prelim test: 167
Number of HSP's that attempted gapping in prelim test: 1929
Number of HSP's gapped (non-prelim): 1636
length of query: 930
length of database: 9,014,727
effective HSP length: 105
effective length of query: 825
effective length of database: 5,132,247
effective search space: 4234103775
effective search space used: 4234103775
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0320b.5


