

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
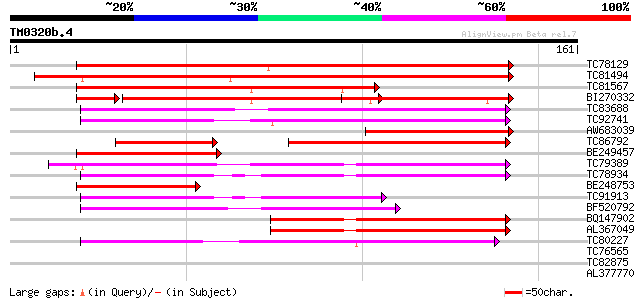
Query= TM0320b.4
(161 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78129 similar to PIR|T47764|T47764 responce reactor 4 - Arabid... 167 2e-42
TC81494 similar to GP|13173408|gb|AAK14395.1 response regulator ... 162 7e-41
TC81567 similar to PIR|T47764|T47764 responce reactor 4 - Arabid... 92 1e-19
BI270332 similar to PIR|T06673|T066 response reactor 2 [imported... 79 3e-17
TC83688 weakly similar to GP|10281000|dbj|BAB13741. pseudo-respo... 69 6e-13
TC92741 similar to GP|10281006|dbj|BAB13743. pseudo-response reg... 67 3e-12
AW683039 similar to PIR|T50857|T508 response regulator 6 [import... 65 2e-11
TC86792 similar to GP|10281004|dbj|BAB13742. pseudo-response reg... 54 9e-11
BE249457 similar to PIR|T50854|T508 response regulator 7 [import... 62 1e-10
TC79389 similar to GP|14189890|dbj|BAB55874. response regulator ... 54 2e-08
TC78934 similar to GP|14189890|dbj|BAB55874. response regulator ... 54 4e-08
BE248753 homologue to GP|11870065|gb| response regulator 15 {Ara... 52 1e-07
TC91913 similar to GP|10281004|dbj|BAB13742. pseudo-response reg... 46 6e-06
BF520792 similar to GP|15131533|em ethylene receptor {Fragaria x... 43 6e-05
BQ147902 similar to PIR|C96700|C96 protein F12A21.15 [imported] ... 42 8e-05
AL367049 similar to GP|13661174|dbj response regulator 8 {Zea ma... 42 1e-04
TC80227 similar to GP|13537198|dbj|BAB40775. histidine kinase {A... 41 2e-04
TC76565 similar to GP|15810171|gb|AAL06987.1 AT3g16857/MUH15_1 {... 39 0.001
TC82875 similar to GP|15131529|emb|CAC48384. ethylene receptor {... 36 0.008
AL377770 similar to PIR|T48853|T488 response reactor 3 [imported... 32 0.086
>TC78129 similar to PIR|T47764|T47764 responce reactor 4 - Arabidopsis
thaliana, partial (62%)
Length = 1307
Score = 167 bits (423), Expect = 2e-42
Identities = 81/133 (60%), Positives = 107/133 (79%), Gaps = 9/133 (6%)
Frame = +1
Query: 20 HVLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRS------- 72
HVLAVDD++IDRKL+E+LL+ SS +VTT ++G +ALEFLGL ++ N S
Sbjct: 406 HVLAVDDSIIDRKLIERLLKTSSYQVTTVDSGSKALEFLGLCENDETNPNTPSVFPNNHQ 585
Query: 73 --KVNLIITDYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFI 130
+VNL+ITDYCMPGMTGY+LLKKIKESS+++ +PVVIMSSEN+P+RIN+CLEEGA+ F
Sbjct: 586 EVEVNLVITDYCMPGMTGYDLLKKIKESSSLRNIPVVIMSSENVPSRINRCLEEGAEEFF 765
Query: 131 LKPLKQSDIKKLK 143
LKP++ SD+ +LK
Sbjct: 766 LKPVRLSDLNRLK 804
>TC81494 similar to GP|13173408|gb|AAK14395.1 response regulator protein
{Dianthus caryophyllus}, partial (53%)
Length = 793
Score = 162 bits (409), Expect = 7e-41
Identities = 85/149 (57%), Positives = 111/149 (74%), Gaps = 13/149 (8%)
Frame = +1
Query: 8 SSNWVMESGDIP---------HVLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFL 58
SS VME+ + HVLAVDD+L+DRK++E+LL+ S+CKVT ++G RAL+FL
Sbjct: 28 SSKRVMETNSVVSIGVDSQEVHVLAVDDSLVDRKVIERLLKISACKVTAVDSGIRALQFL 207
Query: 59 GLT----SGEQNTLNGRSKVNLIITDYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENI 114
GL + E + KV+L+ITDYCMPGMTGYELLKKIKES+ + +PVVIMSSENI
Sbjct: 208 GLVDEKETSESDAFVPGLKVDLVITDYCMPGMTGYELLKKIKESTTFRAIPVVIMSSENI 387
Query: 115 PTRINKCLEEGAQMFILKPLKQSDIKKLK 143
RI++CLEEGA+ FI+KP+K SD+K+LK
Sbjct: 388 LPRIDRCLEEGAEDFIVKPVKLSDVKRLK 474
>TC81567 similar to PIR|T47764|T47764 responce reactor 4 - Arabidopsis
thaliana, partial (27%)
Length = 470
Score = 91.7 bits (226), Expect = 1e-19
Identities = 51/106 (48%), Positives = 70/106 (65%), Gaps = 20/106 (18%)
Frame = +3
Query: 20 HVLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNT------------ 67
HVLAVDD++IDR L+E+LLR+SSCKVT ++G +AL++LGL E ++
Sbjct: 102 HVLAVDDSVIDRMLLERLLRDSSCKVTCVDSGDKALKYLGLNIDEISSTKSSILESSSPS 281
Query: 68 ------LNGRSKVNLIITDYCMPGMTGYELLK--KIKESSAMKEVP 105
L +KVNL +TDYCMPGM+GY+LLK K+K SS + +P
Sbjct: 282 LPQPLQLQEGNKVNLTMTDYCMPGMSGYDLLKRVKVKISSFPRHLP 419
>BI270332 similar to PIR|T06673|T066 response reactor 2 [imported] -
Arabidopsis thaliana, partial (47%)
Length = 556
Score = 79.0 bits (193), Expect(2) = 3e-17
Identities = 44/92 (47%), Positives = 61/92 (65%), Gaps = 18/92 (19%)
Frame = +2
Query: 33 LVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNT------------------LNGRSKV 74
L+E+LLR+SSCKVT ++G +AL++LGL E ++ L +KV
Sbjct: 101 LLERLLRDSSCKVTCVDSGDKALKYLGLNIDEISSTKSSILESSSPSLPQPLQLQEGNKV 280
Query: 75 NLIITDYCMPGMTGYELLKKIKESSAMKEVPV 106
NLI+TDYCMPGM+GY+LLK++K SS K+VPV
Sbjct: 281 NLIMTDYCMPGMSGYDLLKRVKGSS-WKDVPV 373
Score = 42.4 bits (98), Expect = 8e-05
Identities = 24/52 (46%), Positives = 35/52 (67%), Gaps = 3/52 (5%)
Frame = +3
Query: 95 IKESSAM--KEVPVVIMSSENIPTRINKCLEEGAQMFILKPL-KQSDIKKLK 143
+KES + K V+MSSEN+P+RI+ CLEEGA+ F + L D++KL+
Sbjct: 333 LKESRDLLGKMFQFVVMSSENVPSRISMCLEEGAEEFXNEALFSYXDLQKLQ 488
Score = 25.0 bits (53), Expect(2) = 3e-17
Identities = 10/12 (83%), Positives = 12/12 (99%)
Frame = +1
Query: 20 HVLAVDDNLIDR 31
HVLAVDD++IDR
Sbjct: 61 HVLAVDDSVIDR 96
>TC83688 weakly similar to GP|10281000|dbj|BAB13741. pseudo-response
regulator 9 {Arabidopsis thaliana}, partial (27%)
Length = 687
Score = 69.3 bits (168), Expect = 6e-13
Identities = 36/122 (29%), Positives = 69/122 (56%)
Frame = +3
Query: 21 VLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRSKVNLIITD 80
VL V+ + R+L+ LL N KV NG +A E + + + + V+L++T+
Sbjct: 78 VLLVESDRATRRLITSLLNNCHYKVIAVSNGSKAWEMMKMKAID---------VDLVLTE 230
Query: 81 YCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPLKQSDIK 140
+P ++G+ LL I E + +PV++MSS + + + KC+ GA F++KP++++++
Sbjct: 231 MELPAISGFALLSLIMEHEIGRNIPVIMMSSRDSRSTVMKCMCRGAADFLIKPVRKNELT 410
Query: 141 KL 142
L
Sbjct: 411 NL 416
>TC92741 similar to GP|10281006|dbj|BAB13743. pseudo-response regulator 5
{Arabidopsis thaliana}, partial (19%)
Length = 645
Score = 67.0 bits (162), Expect = 3e-12
Identities = 38/123 (30%), Positives = 70/123 (56%), Gaps = 1/123 (0%)
Frame = +3
Query: 21 VLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRSK-VNLIIT 79
VL V+ + R+++ LLR + KV +G +A E L GR + ++LI+T
Sbjct: 243 VLLVEADDCTRQIITALLRKCNYKVAAVADGLKAWEIL----------KGRPRSIDLILT 392
Query: 80 DYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPLKQSDI 139
+ +P ++GY LL I E K +PV++MSS++ + + KC+ GA +++KP+ +++
Sbjct: 393 EVDLPAISGYALLTLIMEHDICKSIPVIMMSSQDSVSTVYKCMLRGAADYLVKPIXINEL 572
Query: 140 KKL 142
+ L
Sbjct: 573 RNL 581
>AW683039 similar to PIR|T50857|T508 response regulator 6 [imported] -
Arabidopsis thaliana, partial (23%)
Length = 384
Score = 64.7 bits (156), Expect = 2e-11
Identities = 28/42 (66%), Positives = 39/42 (92%)
Frame = +1
Query: 102 KEVPVVIMSSENIPTRINKCLEEGAQMFILKPLKQSDIKKLK 143
+E+PVVIMSSEN+ TRI++CLEEGA+ F+LKP+K SD+++LK
Sbjct: 7 REIPVVIMSSENVLTRIDRCLEEGAEDFLLKPVKLSDVRRLK 132
>TC86792 similar to GP|10281004|dbj|BAB13742. pseudo-response regulator 7
{Arabidopsis thaliana}, partial (32%)
Length = 2209
Score = 54.3 bits (129), Expect(2) = 9e-11
Identities = 25/63 (39%), Positives = 41/63 (64%)
Frame = +3
Query: 80 DYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPLKQSDI 139
+ MPG++G LL KI K VPV++MSS + + KCL +GA F++KP++++++
Sbjct: 123 EVAMPGLSGIALLYKIMGHPTRKNVPVIMMSSHDSMGLVFKCLSKGAVDFLVKPIRKNEL 302
Query: 140 KKL 142
K L
Sbjct: 303 KNL 311
Score = 27.7 bits (60), Expect(2) = 9e-11
Identities = 15/29 (51%), Positives = 18/29 (61%)
Frame = +1
Query: 31 RKLVEKLLRNSSCKVTTAENGPRALEFLG 59
R +V LLRN S +V A NG +ALE G
Sbjct: 1 RHIVTALLRNCSYEVIEAANGLQALEGFG 87
>BE249457 similar to PIR|T50854|T508 response regulator 7 [imported] -
Arabidopsis thaliana, partial (21%)
Length = 246
Score = 61.6 bits (148), Expect = 1e-10
Identities = 28/41 (68%), Positives = 36/41 (87%)
Frame = +2
Query: 20 HVLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGL 60
HVLAVDD+L+DRK++E+LL+ SSCKVT E+ RAL+FLGL
Sbjct: 116 HVLAVDDSLVDRKVIERLLKISSCKVTVVESARRALQFLGL 238
>TC79389 similar to GP|14189890|dbj|BAB55874. response regulator 9 {Zea
mays}, partial (33%)
Length = 1532
Score = 54.3 bits (129), Expect = 2e-08
Identities = 40/135 (29%), Positives = 69/135 (50%), Gaps = 4/135 (2%)
Frame = +1
Query: 12 VMESGD-IP---HVLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNT 67
+++SGD P VLAVDD+ ++E LLR VTT AL L
Sbjct: 172 MVDSGDRFPIGMRVLAVDDDPTCLLVLETLLRRCQYHVTTTSQAITALTMLR-------- 327
Query: 68 LNGRSKVNLIITDYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQ 127
+ K +L+I+D MP M G++LL+ + ++PV+++S+ + K + GA
Sbjct: 328 -ENKDKFDLVISDVHMPDMDGFKLLELV---GLEMDLPVIMLSAYGDTKLVMKGISHGAC 495
Query: 128 MFILKPLKQSDIKKL 142
++LKP++ ++K +
Sbjct: 496 DYLLKPVRLEELKNI 540
>TC78934 similar to GP|14189890|dbj|BAB55874. response regulator 9 {Zea
mays}, partial (25%)
Length = 1014
Score = 53.5 bits (127), Expect = 4e-08
Identities = 36/122 (29%), Positives = 63/122 (51%)
Frame = +2
Query: 21 VLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRSKVNLIITD 80
VLAVDD+ L+E LLR+ VTT AL L ++N +L+I +
Sbjct: 68 VLAVDDDPTSLLLLETLLRSCQYHVTTTSEAITALTML-----QENI----DMFDLVIAE 220
Query: 81 YCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPLKQSDIK 140
MP M G +LL+ + ++PV+++S+ + K + GA+ F+LKP++ +++
Sbjct: 221 VHMPDMDGLKLLELV---GLEMDLPVIMLSAHGETELVMKAISHGARDFLLKPVRLEELR 391
Query: 141 KL 142
+
Sbjct: 392 NI 397
>BE248753 homologue to GP|11870065|gb| response regulator 15 {Arabidopsis
thaliana}, partial (22%)
Length = 257
Score = 51.6 bits (122), Expect = 1e-07
Identities = 22/35 (62%), Positives = 30/35 (84%)
Frame = +2
Query: 20 HVLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRA 54
HVLAVDD+ +DRK++E+LL+ +SCKVT E+G RA
Sbjct: 122 HVLAVDDSFVDRKVIERLLKFTSCKVTVVESGTRA 226
>TC91913 similar to GP|10281004|dbj|BAB13742. pseudo-response regulator 7
{Arabidopsis thaliana}, partial (13%)
Length = 907
Score = 46.2 bits (108), Expect = 6e-06
Identities = 33/87 (37%), Positives = 47/87 (53%)
Frame = +1
Query: 21 VLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRSKVNLIITD 80
VL V+ + R +V LLRN S +V A NG +A + L E T + ++LI+T+
Sbjct: 538 VLLVEYDDCTRHIVTALLRNCSYEVIEAANGLQAWKVL-----EDLT----NHIDLILTE 690
Query: 81 YCMPGMTGYELLKKIKESSAMKEVPVV 107
MPG++G LL KI K VPV+
Sbjct: 691 VAMPGLSGIALLYKIMGHPTRKNVPVI 771
>BF520792 similar to GP|15131533|em ethylene receptor {Fragaria x ananassa},
partial (22%)
Length = 619
Score = 42.7 bits (99), Expect = 6e-05
Identities = 24/91 (26%), Positives = 47/91 (51%)
Frame = +1
Query: 21 VLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRSKVNLIITD 80
VL VD++ ++R + +KLL+ C VT+A G L +G S + +I+ D
Sbjct: 349 VLLVDNDDVNRAVTQKLLQKLGCSVTSASTGFECLTVIGPVG---------SSIQVILLD 501
Query: 81 YCMPGMTGYELLKKIKESSAMKEVPVVIMSS 111
MP + G+E+ +I++ + +V +++
Sbjct: 502 LQMPDIDGFEVAARIRKFKSGNRPIIVALTA 594
>BQ147902 similar to PIR|C96700|C96 protein F12A21.15 [imported] -
Arabidopsis thaliana, partial (19%)
Length = 363
Score = 42.4 bits (98), Expect = 8e-05
Identities = 18/68 (26%), Positives = 43/68 (62%)
Frame = +1
Query: 75 NLIITDYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPL 134
+++I+D P M G++LL+ + ++PV++MS + +R+ K ++ GA ++LKP+
Sbjct: 100 DIVISDVNTPDMDGFKLLEHV---GLEMDLPVIMMSVDGETSRVMKGVQHGACDYLLKPI 270
Query: 135 KQSDIKKL 142
+ +++ +
Sbjct: 271 RMKEVRNI 294
>AL367049 similar to GP|13661174|dbj response regulator 8 {Zea mays}, partial
(13%)
Length = 283
Score = 41.6 bits (96), Expect = 1e-04
Identities = 18/68 (26%), Positives = 42/68 (61%)
Frame = +2
Query: 75 NLIITDYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPL 134
+++++D MP M GY+LL+ + ++PV++MS ++ + + K + GA F +KP+
Sbjct: 59 DVVLSDVHMPDMDGYKLLEHV---GLEMDLPVIMMSVDDRTSTVMKGIRHGACDFFIKPV 229
Query: 135 KQSDIKKL 142
+ +++ +
Sbjct: 230 RMEELRNI 253
>TC80227 similar to GP|13537198|dbj|BAB40775. histidine kinase {Arabidopsis
thaliana}, partial (22%)
Length = 1693
Score = 41.2 bits (95), Expect = 2e-04
Identities = 30/127 (23%), Positives = 54/127 (41%), Gaps = 8/127 (6%)
Frame = +3
Query: 21 VLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRSKVNLIITD 80
+L VDDN ++R + L+ V +G A+ ++L + + D
Sbjct: 768 ILIVDDNGVNRAVAAGALKKYGADVVCVSSGKDAI----------SSLKPPHQFDACFMD 917
Query: 81 YCMPGMTGYELLKKIKE--------SSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILK 132
MP M G+E +I+E +S VP++ M+++ I KCL G ++ K
Sbjct: 918 IQMPEMDGFEATGRIREMEQNVNREASINWHVPILAMTADVIQATHEKCLRCGMDGYVSK 1097
Query: 133 PLKQSDI 139
P + +
Sbjct: 1098PFEAEQL 1118
>TC76565 similar to GP|15810171|gb|AAL06987.1 AT3g16857/MUH15_1 {Arabidopsis
thaliana}, partial (31%)
Length = 842
Score = 38.9 bits (89), Expect = 0.001
Identities = 19/68 (27%), Positives = 43/68 (62%)
Frame = +2
Query: 75 NLIITDYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPL 134
+++I+D MP M G++LL+ I ++PV++MS+++ + + K + GA +++KP+
Sbjct: 41 DIVISDVHMPDMDGFKLLEHI---GLEMDLPVIMMSADDGKSVVMKGVTHGACDYLIKPV 211
Query: 135 KQSDIKKL 142
+ +K +
Sbjct: 212 RIEALKNI 235
>TC82875 similar to GP|15131529|emb|CAC48384. ethylene receptor {Fragaria x
ananassa}, partial (39%)
Length = 1118
Score = 35.8 bits (81), Expect = 0.008
Identities = 27/109 (24%), Positives = 47/109 (42%), Gaps = 2/109 (1%)
Frame = +3
Query: 21 VLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRSKVNLIITD 80
VL +DDN + R + LL + C VTT + L + L + ++ D
Sbjct: 543 VLVMDDNGVSRSATKGLLIHLGCDVTTVSSSEECLRVVSL------------EHKVVFMD 686
Query: 81 YCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINK--CLEEGAQ 127
C + GYEL +I+E ++ +I++ ++ K CL G +
Sbjct: 687 VC-ARLDGYELAVRIQEKFTKRQDRPLIVALTGNTNKLTKENCLRAGVK 830
>AL377770 similar to PIR|T48853|T488 response reactor 3 [imported] -
Arabidopsis thaliana, partial (12%)
Length = 487
Score = 32.3 bits (72), Expect = 0.086
Identities = 13/22 (59%), Positives = 20/22 (90%)
Frame = +3
Query: 122 LEEGAQMFILKPLKQSDIKKLK 143
LEEGA+ F++KPL+ SD++KL+
Sbjct: 3 LEEGAEEFLMKPLQLSDLQKLQ 68
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.132 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,874,180
Number of Sequences: 36976
Number of extensions: 56379
Number of successful extensions: 309
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 303
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 303
length of query: 161
length of database: 9,014,727
effective HSP length: 88
effective length of query: 73
effective length of database: 5,760,839
effective search space: 420541247
effective search space used: 420541247
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0320b.4


