

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0318.6
(172 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
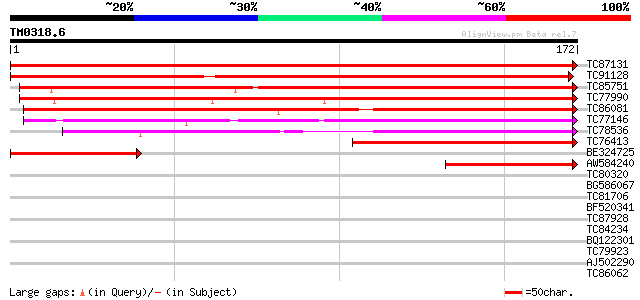
Score E
Sequences producing significant alignments: (bits) Value
TC87131 weakly similar to GP|5031281|gb|AAD38146.1| unknown {Pru... 289 3e-79
TC91128 similar to GP|5031281|gb|AAD38146.1| unknown {Prunus arm... 278 6e-76
TC85751 similar to GP|5031281|gb|AAD38146.1| unknown {Prunus arm... 239 4e-64
TC77990 similar to GP|5031281|gb|AAD38146.1| unknown {Prunus arm... 218 1e-57
TC86081 similar to GP|4510345|gb|AAD21434.1| expressed protein {... 161 1e-40
TC77146 weakly similar to PIR|D84674|D84674 hypothetical protein... 143 4e-35
TC78536 similar to GP|21536983|gb|AAM61324.1 unknown {Arabidopsi... 109 5e-25
TC76413 similar to GP|3461820|gb|AAC32914.1| unknown protein {Ar... 101 1e-22
BE324725 similar to GP|5031281|gb|A unknown {Prunus armeniaca}, ... 88 1e-18
AW584240 46 6e-06
TC80320 similar to GP|21439798|emb|CAD35172. unnamed protein pro... 30 0.48
BG586067 similar to GP|21592847|gb unknown {Arabidopsis thaliana... 29 0.82
TC81706 similar to PIR|T50672|T50672 probable zinc finger protei... 28 1.8
BF520341 similar to GP|23318573|gb Sequence 89 from patent US 64... 28 2.4
TC87928 similar to PIR|A96839|A96839 F23A5.2(form2) [imported] -... 27 4.1
TC84234 similar to PIR|D96586|D96586 hypothetical protein F20D21... 27 4.1
BQ122301 similar to PIR|T51496|T514 hypothetical protein T21H19_... 27 5.3
TC79923 similar to PIR|T04748|T04748 hypothetical protein T16H5.... 27 5.3
AJ502290 similar to PIR|T11021|T11 farnesyltranstransferase (EC ... 27 5.3
TC86062 similar to GP|21592847|gb|AAM64797.1 unknown {Arabidopsi... 27 5.3
>TC87131 weakly similar to GP|5031281|gb|AAD38146.1| unknown {Prunus
armeniaca}, partial (82%)
Length = 982
Score = 289 bits (740), Expect = 3e-79
Identities = 135/172 (78%), Positives = 151/172 (87%)
Frame = +2
Query: 1 MESHDETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVN 60
MESHDE GCQAPERPILCVNNCGFFGR ATMNMCSKCYKD LLKQEQ+KL ATSVENIVN
Sbjct: 230 MESHDEMGCQAPERPILCVNNCGFFGREATMNMCSKCYKDTLLKQEQEKLVATSVENIVN 409
Query: 61 SCSNGNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRV 120
S+ NGK A+TA AV+VRVE VE+ V+ ++ + S ES+E+KAKTGPSRC TCRKRV
Sbjct: 410 GNSSSNGKLAVTASAVDVRVESVELNTVSPEVPENPISNESVEMKAKTGPSRCATCRKRV 589
Query: 121 GLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
GLTGFSCKCGN+FC+MHRYSDKHDCPFDYR GQ+AIA++NPVIKADKLDKI
Sbjct: 590 GLTGFSCKCGNLFCSMHRYSDKHDCPFDYRTAGQKAIAESNPVIKADKLDKI 745
>TC91128 similar to GP|5031281|gb|AAD38146.1| unknown {Prunus armeniaca},
partial (64%)
Length = 747
Score = 278 bits (712), Expect = 6e-76
Identities = 131/171 (76%), Positives = 150/171 (87%)
Frame = +1
Query: 1 MESHDETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVN 60
M+SHD+TGCQ P+ PILCVNNCGFFGRAATMNMCSKCYKD L QEQ+KLAA SVEN+V
Sbjct: 121 MDSHDKTGCQTPKLPILCVNNCGFFGRAATMNMCSKCYKDTQLMQEQEKLAAASVENLV- 297
Query: 61 SCSNGNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRV 120
S G+ KQ +T AVNV++E VEVK V+A+IS DSSS E+LE K KTGPSRC TCRKRV
Sbjct: 298 --SGGSMKQVVTDGAVNVQIENVEVKTVSAEISGDSSSSENLETKVKTGPSRCATCRKRV 471
Query: 121 GLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDK 171
GLTGF+CKCGN+FCAMHRYSDKHDCPFDY++VG++AIAK+NPVIKADKLDK
Sbjct: 472 GLTGFTCKCGNLFCAMHRYSDKHDCPFDYQSVGRDAIAKSNPVIKADKLDK 624
>TC85751 similar to GP|5031281|gb|AAD38146.1| unknown {Prunus armeniaca},
partial (77%)
Length = 1128
Score = 239 bits (610), Expect = 4e-64
Identities = 113/171 (66%), Positives = 140/171 (81%), Gaps = 2/171 (1%)
Frame = +1
Query: 4 HDETGCQA-PERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSC 62
HD+TGC+A PE P+LC+NNCGFFG AATMNMCSKC+KDM+LKQEQ LAA+S+ NI+N
Sbjct: 217 HDQTGCEAAPEGPMLCINNCGFFGSAATMNMCSKCHKDMMLKQEQATLAASSIGNIMNGS 396
Query: 63 SNGNG-KQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRVG 121
S+ +G + AITA+ V + V+PVE K ++A+ S S ESLE K K GP RC C KRVG
Sbjct: 397 SSSSGIEPAITAN-VEISVDPVEPKIISAEPLVASGSEESLEKKPKDGPKRCSNCNKRVG 573
Query: 122 LTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
LTGF+C+CGN++CA+HRYSDKHDCPFDYR G++AIAKANPV+KA+KLDKI
Sbjct: 574 LTGFNCRCGNLYCAVHRYSDKHDCPFDYRTAGRDAIAKANPVVKAEKLDKI 726
>TC77990 similar to GP|5031281|gb|AAD38146.1| unknown {Prunus armeniaca},
partial (79%)
Length = 1194
Score = 218 bits (554), Expect = 1e-57
Identities = 109/185 (58%), Positives = 133/185 (70%), Gaps = 16/185 (8%)
Frame = +2
Query: 4 HDETGCQAP-ERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVN-S 61
H++T CQAP E PILC+NNCGFFG AATMNMCSKC+KDM+LKQEQ +LAA+S+ NI+N S
Sbjct: 272 HEKTECQAPPEGPILCINNCGFFGSAATMNMCSKCHKDMMLKQEQAQLAASSLGNIMNGS 451
Query: 62 CSNGNGKQAITADAVNVRVEPVEVKAVTAQISA--------------DSSSGESLEVKAK 107
SN + +TA +V++ VE K + I A S S ES + K K
Sbjct: 452 TSNTEKEPVVTATSVDIPAISVEPKTASVDIPAISVEPETISKPFLFGSGSEESDDPKPK 631
Query: 108 TGPSRCGTCRKRVGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKAD 167
GP RC C KRVGLTGF+C+CGN+FCA+HRYSDKHDCPFDYR ++AIAKANPV+KA+
Sbjct: 632 DGPKRCSNCNKRVGLTGFNCRCGNLFCAVHRYSDKHDCPFDYRTSARDAIAKANPVVKAE 811
Query: 168 KLDKI 172
KLDKI
Sbjct: 812 KLDKI 826
>TC86081 similar to GP|4510345|gb|AAD21434.1| expressed protein {Arabidopsis
thaliana}, partial (72%)
Length = 1066
Score = 161 bits (408), Expect = 1e-40
Identities = 77/170 (45%), Positives = 106/170 (62%), Gaps = 2/170 (1%)
Frame = +3
Query: 5 DETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSCSN 64
+E C+ PE LC NNCGFFG +ATMN+CSKCY+D+ LK+++ +++E ++S S
Sbjct: 195 EEHRCETPEGHRLCANNCGFFGSSATMNLCSKCYRDIHLKEQEQAKTKSTIETALSSASA 374
Query: 65 GNGKQAITADAVNVRV--EPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRVGL 122
+ + +P I ++S S V++ +RCGTCRKR GL
Sbjct: 375 STAVVVAASPVAEIESLPQPQPPALTVPSIVPEASDNSSGPVQS----NRCGTCRKRTGL 542
Query: 123 TGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
TGF C+CG FC HRY +KH+C FD++AVG+E IAKANPVIKADKL +I
Sbjct: 543 TGFKCRCGITFCGSHRYPEKHECGFDFKAVGREEIAKANPVIKADKLRRI 692
>TC77146 weakly similar to PIR|D84674|D84674 hypothetical protein At2g27580
[imported] - Arabidopsis thaliana, partial (74%)
Length = 956
Score = 143 bits (360), Expect = 4e-35
Identities = 74/173 (42%), Positives = 104/173 (59%), Gaps = 5/173 (2%)
Frame = +3
Query: 5 DETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAA-----TSVENIV 59
+E CQA +R LC NNCGFFG A ++CSKCY+D+ +K+++ A T +
Sbjct: 267 EEHRCQAAQR--LCANNCGFFGSPAMQDLCSKCYRDLQMKEQRSSSAKLVLNQTLIPQQS 440
Query: 60 NSCSNGNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKR 119
NS S G I + + V V T ++ A +++G S K P+RCGTCR+R
Sbjct: 441 NSSSLDTG--IIHPSSTSPSVMIVSSSTPTVELVA-AAAGPSEAEPPKVQPNRCGTCRRR 611
Query: 120 VGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
VGLTGF C+CG C HRY ++H C FD++ +G+E I KANPV+K +KL+KI
Sbjct: 612 VGLTGFKCRCGLTLCGTHRYPEQHGCGFDFKGMGREEIKKANPVVKGEKLNKI 770
>TC78536 similar to GP|21536983|gb|AAM61324.1 unknown {Arabidopsis
thaliana}, partial (48%)
Length = 972
Score = 109 bits (273), Expect = 5e-25
Identities = 58/158 (36%), Positives = 82/158 (51%), Gaps = 2/158 (1%)
Frame = +2
Query: 17 LCVNNCGFFGRAATMNMCSKCY--KDMLLKQEQDKLAATSVENIVNSCSNGNGKQAITAD 74
LC+ NCG G +T NMC C+ +L + + + S S+ + + D
Sbjct: 209 LCIKNCGVVGNPSTNNMCQNCFTASTTILPPSSSRSVRSPKRSRQESSSSSEEEGSTDHD 388
Query: 75 AVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRVGLTGFSCKCGNVFC 134
V+ + EVK V SRC CR++VGL GF C+CG +FC
Sbjct: 389 LVDEKTVS-EVKRVV---------------------SRCSGCRRKVGLAGFRCRCGELFC 502
Query: 135 AMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
A HRYSD+HDC +DY+ VG+E IA+ NPVI+A K+ K+
Sbjct: 503 ADHRYSDRHDCGYDYKKVGREEIARENPVIRAAKIVKV 616
>TC76413 similar to GP|3461820|gb|AAC32914.1| unknown protein {Arabidopsis
thaliana}, partial (54%)
Length = 1310
Score = 101 bits (252), Expect = 1e-22
Identities = 41/68 (60%), Positives = 56/68 (82%)
Frame = +3
Query: 105 KAKTGPSRCGTCRKRVGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVI 164
+AK +RC CRKRVGLTGF C+CG++FC+ HRYSD+HDC +DY+A G+E+IA+ NPV+
Sbjct: 405 EAKRAVNRCSGCRKRVGLTGFRCRCGDLFCSEHRYSDRHDCSYDYKAAGRESIARENPVV 584
Query: 165 KADKLDKI 172
KA K+ K+
Sbjct: 585 KAAKIVKL 608
>BE324725 similar to GP|5031281|gb|A unknown {Prunus armeniaca}, partial
(16%)
Length = 499
Score = 88.2 bits (217), Expect = 1e-18
Identities = 37/40 (92%), Positives = 38/40 (94%)
Frame = +3
Query: 1 MESHDETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKD 40
M+SHDETGCQ PE PILCVNNCGFFGRAATMNMCSKCYKD
Sbjct: 282 MDSHDETGCQTPELPILCVNNCGFFGRAATMNMCSKCYKD 401
>AW584240
Length = 391
Score = 46.2 bits (108), Expect = 6e-06
Identities = 19/40 (47%), Positives = 27/40 (67%)
Frame = -3
Query: 133 FCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
FC R +K +C FD++ VG++ I K NPVIK DK+ +I
Sbjct: 383 FCGSPRDPEKQECGFDFKTVGKKEITKTNPVIKGDKVRRI 264
>TC80320 similar to GP|21439798|emb|CAD35172. unnamed protein product {Zea
mays}, partial (92%)
Length = 709
Score = 30.0 bits (66), Expect = 0.48
Identities = 14/50 (28%), Positives = 20/50 (40%), Gaps = 9/50 (18%)
Frame = +2
Query: 9 CQAPERPILCVNNCGFF---------GRAATMNMCSKCYKDMLLKQEQDK 49
C RP +C+ CG G MC +CY DM+ + + K
Sbjct: 317 CSVHSRPNVCMRACGTCCLRCKCVPPGTYGNREMCGRCYTDMITRGNKPK 466
>BG586067 similar to GP|21592847|gb unknown {Arabidopsis thaliana}, partial
(82%)
Length = 762
Score = 29.3 bits (64), Expect = 0.82
Identities = 15/44 (34%), Positives = 23/44 (52%)
Frame = +3
Query: 92 ISADSSSGESLEVKAKTGPSRCGTCRKRVGLTGFSCKCGNVFCA 135
+S +SSS + G S G C +R+G+ F C G+ +CA
Sbjct: 99 VSRNSSSDTTSRF*NNGGRSGFGYCNQRLGVNPFICCYGSYWCA 230
>TC81706 similar to PIR|T50672|T50672 probable zinc finger protein
[imported] - Arabidopsis thaliana, partial (67%)
Length = 780
Score = 28.1 bits (61), Expect = 1.8
Identities = 11/23 (47%), Positives = 13/23 (55%), Gaps = 1/23 (4%)
Frame = +3
Query: 125 FSCK-CGNVFCAMHRYSDKHDCP 146
F+C C VFC HR H+CP
Sbjct: 141 FTCDGCKQVFCVEHRSYKSHECP 209
>BF520341 similar to GP|23318573|gb Sequence 89 from patent US 6417428,
partial (68%)
Length = 645
Score = 27.7 bits (60), Expect = 2.4
Identities = 8/22 (36%), Positives = 12/22 (54%)
Frame = -3
Query: 114 GTCRKRVGLTGFSCKCGNVFCA 135
G C+ G+ C CGN+ C+
Sbjct: 379 GVCKVETGVAAPQCHCGNIMCS 314
>TC87928 similar to PIR|A96839|A96839 F23A5.2(form2) [imported] -
Arabidopsis thaliana, partial (95%)
Length = 1519
Score = 26.9 bits (58), Expect = 4.1
Identities = 12/28 (42%), Positives = 15/28 (52%), Gaps = 1/28 (3%)
Frame = +1
Query: 129 CGNVFCAMHRYS-DKHDCPFDYRAVGQE 155
C + C RY D D PF +R +GQE
Sbjct: 493 CSHARCTYQRYCLDSRDEPFSHRKLGQE 576
>TC84234 similar to PIR|D96586|D96586 hypothetical protein F20D21.27
[imported] - Arabidopsis thaliana, partial (19%)
Length = 721
Score = 26.9 bits (58), Expect = 4.1
Identities = 28/93 (30%), Positives = 40/93 (42%), Gaps = 1/93 (1%)
Frame = +1
Query: 29 ATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSCSNGNGKQAITADAVNVRVEPVEVKAV 88
A +N K ++ L E ++L TS+ N S + N ++ P A
Sbjct: 421 ANLNFVEKLFEQWLPLPESNRLV-TSLLNDAKSGAPLNVPGNCSSPNAATNSLPSMFPAG 597
Query: 89 TAQ-ISADSSSGESLEVKAKTGPSRCGTCRKRV 120
TA +S SSSG VK + GPS G+ K V
Sbjct: 598 TAPPLSPRSSSGSPRIVKQRAGPSNLGSPLKVV 696
>BQ122301 similar to PIR|T51496|T514 hypothetical protein T21H19_180 -
Arabidopsis thaliana, partial (5%)
Length = 511
Score = 26.6 bits (57), Expect = 5.3
Identities = 15/42 (35%), Positives = 19/42 (44%), Gaps = 4/42 (9%)
Frame = +3
Query: 122 LTGFSCKCGNVFCAMHRYSDKHDCP----FDYRAVGQEAIAK 159
L FSC + MH Y+ H CP + YR Q +I K
Sbjct: 285 LLNFSCVLYTIITPMHCYTCDHSCPSFWIWLYRLQKQTSIRK 410
>TC79923 similar to PIR|T04748|T04748 hypothetical protein T16H5.30 -
Arabidopsis thaliana, partial (28%)
Length = 1001
Score = 26.6 bits (57), Expect = 5.3
Identities = 14/43 (32%), Positives = 20/43 (45%), Gaps = 4/43 (9%)
Frame = +2
Query: 112 RCGTCRKRVGLTG----FSCKCGNVFCAMHRYSDKHDCPFDYR 150
RC CR + LT +C+CG+ FC + C +YR
Sbjct: 779 RCQQCRIMIELTQGCYHMTCRCGHEFC--------YSCGAEYR 883
>AJ502290 similar to PIR|T11021|T11 farnesyltranstransferase (EC 2.5.1.29) -
white lupine, partial (41%)
Length = 657
Score = 26.6 bits (57), Expect = 5.3
Identities = 10/29 (34%), Positives = 16/29 (54%)
Frame = +2
Query: 14 RPILCVNNCGFFGRAATMNMCSKCYKDML 42
RP+LC+ CG G ++ S C +M+
Sbjct: 395 RPVLCIAACGLVGGKEEASIPSACAVEMI 481
>TC86062 similar to GP|21592847|gb|AAM64797.1 unknown {Arabidopsis
thaliana}, partial (97%)
Length = 1261
Score = 26.6 bits (57), Expect = 5.3
Identities = 11/27 (40%), Positives = 16/27 (58%)
Frame = +3
Query: 109 GPSRCGTCRKRVGLTGFSCKCGNVFCA 135
G S G C +R+G+ F C G+ +CA
Sbjct: 204 GRSGFGYCNQRLGVNPFICCYGSYWCA 284
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.131 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,897,610
Number of Sequences: 36976
Number of extensions: 82931
Number of successful extensions: 402
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 393
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 396
length of query: 172
length of database: 9,014,727
effective HSP length: 89
effective length of query: 83
effective length of database: 5,723,863
effective search space: 475080629
effective search space used: 475080629
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0318.6


