

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0318.12
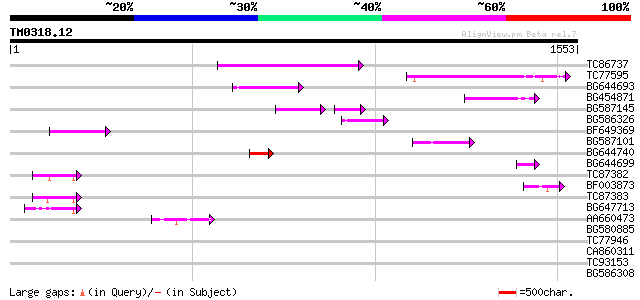
(1553 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 252 7e-67
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 182 7e-46
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 150 5e-36
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 127 3e-29
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 96 8e-26
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 97 6e-20
BF649369 86 1e-16
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 73 1e-12
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 71 3e-12
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 56 9e-08
TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulch... 52 1e-06
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 50 7e-06
TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA PO... 50 9e-06
BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent vir... 49 1e-05
AA660473 44 6e-04
BG580885 40 0.007
TC77946 homologue to GP|10334491|emb|CAC10207. putative splicing... 39 0.020
CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product ... 38 0.027
TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Ci... 38 0.035
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 37 0.046
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 252 bits (644), Expect = 7e-67
Identities = 159/422 (37%), Positives = 234/422 (54%), Gaps = 21/422 (4%)
Frame = +1
Query: 569 GEGYVLQYQMLEKEVAKAEETP-----ELLKAVLSDFPALFAAPT--ELPPQRRH-DHAI 620
G V+ + + E A A + P + VL +FP LF ++P R DHAI
Sbjct: 247 GAYQVMSVSLEDIEKALAPKAPVDPRKHVPAGVLEEFPDLFNPEKAYQVPASRGLLDHAI 426
Query: 621 HL---KEGAS--IPNIRPYRYPHYQKNEIEKLVAEMLNAGIIRPSISPYSSPVILVRKKD 675
L K+G +P Y + ++K + ++L+ G I+ S S +PV+ VRK
Sbjct: 427 PLIPDKDGNDPPLPWGPLYGMSRQELLVLKKTLEDLLDKGFIKASGSAAGAPVLFVRKPG 606
Query: 676 GSWRFCVDYRALNKITVPNKFPIPVIDELLDEIGPARVFSKIDLKSGYHQIMMKAEDVEK 735
G RFCVDYRALN IT +++P+P+I E L + AR F+K+D+ + +H++ +K ED EK
Sbjct: 607 GGIRFCVDYRALNAITKKDRYPLPLISETLRRVAGARWFTKLDVVAAFHKMRIKDEDQEK 786
Query: 736 TAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLRPVLRKCALVFFDDILIYSL-TMEEH 794
TAFRT G +E++V PFGLT AP+TFQ +N+ L L + DD+LIY+ + ++H
Sbjct: 787 TAFRTRYGLFEWIVCPFGLTGAPATFQRYINKTLHEFLDDFVTAYIDDVLIYTTGSKKDH 966
Query: 795 VVHLTQVLQLMDQHDLKINGKKSVFGRGQIEYLGHVLSAGT-VAADPKKLEAMWLWPVPK 853
+ +VL+ + L ++ KK F ++Y+G +L+AG V+ DP KL A+ W P
Sbjct: 967 EAQVRRVLRRLADAGLSLDPKKCEFSVTTVKYVGFILTAGKGVSCDPLKLAAIRDWLPPG 1146
Query: 854 DLKSLRGFLGLTGYYRRFVKDYGKIARPLTQLLKKD-SFQWGPSPQQAFEALKHTLTEIP 912
+K R FLG YY+ F+ Y +I PLT+L +KD F+WG + AF LK E P
Sbjct: 1147SVKGARSFLGFCNYYKDFIPGYSEITEPLTRLTRKDFPFRWGAEQEAAFTKLKRLFAEEP 1326
Query: 913 TLAVPDFSKVFVLETDASGTGLGAVLTQE-----GKPLAFWSATLSDRSQAKSVYERELM 967
L + D V +ETD SG LG VLTQE P+AF S LS ++++EL+
Sbjct: 1327VLRMFDPEAVTTVETDCSGFALGGVLTQEDGTGAAHPVAFHSQRLSPAEYNYPIHDKELL 1506
Query: 968 AV 969
AV
Sbjct: 1507AV 1512
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 182 bits (463), Expect = 7e-46
Identities = 140/471 (29%), Positives = 220/471 (45%), Gaps = 23/471 (4%)
Frame = +2
Query: 1088 RLFYKGKL-VLSKQSSRIPI------LCKEFHASLLGGHSGFFRTYRRLAAVVYWEGMKS 1140
RL ++G++ V P+ L +E H S GH G T ++ +W G
Sbjct: 101 RLTFRGRIWVPGSDDEESPLNELRTKLVQESHDSTAAGHPGRNGTLEIVSRKFFWPGQSQ 280
Query: 1141 DIRDFVAECDTCQRNKFDNLSPAGLLQPLPVPTQVWTDVSMDFIGGLPKSNGKDT--ILV 1198
+R FV CD C + G L+PLPVP ++ +D+SMDFI LP + G+ + + V
Sbjct: 281 TVRRFVRNCDVCGGIHIWRQAKRGFLKPLPVPNRLHSDLSMDFITSLPPTRGRGSQYLWV 460
Query: 1199 VVDRLTKYAHFIPLRHPFTAPEVAALFLHEVVRLHGFPTTIVSDRDSLFLSSFWKELFRV 1258
+VDRL+K + A A FL R HG P +IVSDR S ++ FW+E R+
Sbjct: 461 IVDRLSKSVTLEEM-DTMEAEACAQRFLSCHYRFHGMPQSIVSDRGSNWVGRFWREFCRL 637
Query: 1259 SGTQLKFSSAYHPQTDGQTEVVNRCLEVYLRCLTGSRPRKWVTCLPWAEFWFNSNYNRSA 1318
+G S++YHPQTDG TE N+ ++ LR W LP + + +N S
Sbjct: 638 TGVTQLLSTSYHPQTDGGTERWNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSI 817
Query: 1319 KMSPFQALYGRE----PPVLLQGTTIPSKIAAVNDLQVGRDELLSDLRANLLKSQDMMRA 1374
+PF +G P V G + AA L ++ ++A ++ +Q A
Sbjct: 818 GATPFFVEHGYHVDPIPTVEDTGGVVSEGEAAAQLLVKRMKDVTGFIQAEIVAAQQRSEA 997
Query: 1375 YANKKRRDVD-YQIGDEVFLKLQPYRRRSLAKKMNEKLSPRYYGPYPIVAKIGAVAYRLE 1433
ANK+R D YQ+GD+V+L + Y+ +KK++ + Y + + L
Sbjct: 998 SANKRRCPADRYQVGDKVWLNVSNYKSPRPSKKLD-----WLHHKYEVTRFVTPHVVELN 1162
Query: 1434 LPAHSRVHPVFHVSLLKTAVGTNF------QPQPLPAALNEDHELLVEPESVLAVRETTM 1487
+P V+P FHV LL+ A PQP P +++D E+ E E +LA R +
Sbjct: 1163 VP--GTVYPKFHVDLLRRAASDPLPGQEVVDPQP-PPIVDDDGEVEWEVEEILAARWHQV 1333
Query: 1488 G---QIEVLIQWHSLPACENSWESAAQLHEAFPDFPLEDKVKHLGGGIDEH 1535
G + + L++W + +WE+A +A + D+ + G ID+H
Sbjct: 1334 GRGRRRQALVKWKGF--VDATWEAA----DAIRETEALDRYEARFGPIDDH 1468
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 150 bits (378), Expect = 5e-36
Identities = 84/201 (41%), Positives = 115/201 (56%), Gaps = 5/201 (2%)
Frame = +2
Query: 610 LPPQRRHDHAIHLKEGASIPNIRPYRYPHYQKNEIEKLVA-----EMLNAGIIRPSISPY 664
+PP+ + D I L +PN+ P P Y+ N ++ V ++L G I+PSI P
Sbjct: 17 VPPEWKIDFGIDL-----LPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP* 181
Query: 665 SSPVILVRKKDGSWRFCVDYRALNKITVPNKFPIPVIDELLDEIGPARVFSKIDLKSGYH 724
V+ ++KKDG R +DY LN + + K+P+P+IDEL D + ++ F KIDL+ G H
Sbjct: 182 GVVVLFLKKKDGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*H 361
Query: 725 QIMMKAEDVEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLRPVLRKCALVFFDDI 784
Q + EDV KTAFR GHYE LVM FG TN P F LMN V + L +VF +DI
Sbjct: 362 QHRVIGEDVPKTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDI 541
Query: 785 LIYSLTMEEHVVHLTQVLQLM 805
LIYS EH HL L+++
Sbjct: 542 LIYSKNENEHENHLRLALKVL 604
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 127 bits (319), Expect = 3e-29
Identities = 79/211 (37%), Positives = 115/211 (54%), Gaps = 5/211 (2%)
Frame = +2
Query: 1245 SLFLSSFWKELFRVSGTQLKFSSAYHPQTDGQTEVVNRCLEVYLRCLTGSRPRKWVTCLP 1304
S S+FWK+LF++ GT L SSAYHP +DGQ+E +N+ E+YLRCL + P KW P
Sbjct: 20 SSLYSNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFP 199
Query: 1305 WAEFWFNSNYNRSAKMSPFQALYGREPPVLLQGTTIPSKIAAVNDLQVGRDELLSDL--- 1361
WAE+W+N++YN SA M+PF+ALYGR+ +L++ A + R+ELLS L
Sbjct: 200 WAEYWYNTSYNISAAMTPFKALYGRDLSMLIRSKGSSKDTADLQSQLAQREELLSQLQSI 379
Query: 1362 --RANLLKSQDMMRAYANKKRRDVDYQIGDEVFLKLQPYRRRSLAKKMNEKLSPRYYGPY 1419
R N L S ++R A V + + + R + + P
Sbjct: 380 STRLNKL*SIKLIRNAAILSSSWVSTSL*NCNLIN----SLR*HCGNIKSSVHP------ 529
Query: 1420 PIVAKIGAVAYRLELPAHSRVHPVFHVSLLK 1450
+V + AY+L LP+ ++V P+FHVS LK
Sbjct: 530 TLVHY*QSAAYKLSLPSTAKVPPIFHVSQLK 622
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 96.3 bits (238), Expect(2) = 8e-26
Identities = 49/136 (36%), Positives = 81/136 (59%)
Frame = +2
Query: 728 MKAEDVEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLRPVLRKCALVFFDDILIY 787
M +D+EKTAF T G Y + VMPFGL NA ST+Q L+N + L V+ DD+L+
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 788 SLTMEEHVVHLTQVLQLMDQHDLKINGKKSVFGRGQIEYLGHVLSAGTVAADPKKLEAMW 847
SL +H+ HL + + +D++ +K+N K FG E+LG++++ + +PK++ A+
Sbjct: 182 SLRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAIL 361
Query: 848 LWPVPKDLKSLRGFLG 863
P PK+ + ++ G
Sbjct: 362 DLPSPKNSREVQRLTG 409
Score = 40.8 bits (94), Expect(2) = 8e-26
Identities = 25/89 (28%), Positives = 41/89 (45%), Gaps = 4/89 (4%)
Frame = +3
Query: 891 FQWGPSPQQAFEALKHTLTEIPTLAVPDFSKVFVLETDASGTGLGAVLTQEG----KPLA 946
F W ++AFE LK LT P L+ P+ L S T + +VL +E KP+
Sbjct: 495 FVWDEKCEEAFEQLKQYLTTPPVLSKPEAGDTLSLYIAISSTAVSSVLIREDRGEQKPIF 674
Query: 947 FWSATLSDRSQAKSVYERELMAVVRAVQR 975
+ S ++D E+ AV+ + ++
Sbjct: 675 YTSKRMTDPETRYPTLEKMAFAVITSARK 761
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 96.7 bits (239), Expect = 6e-20
Identities = 55/130 (42%), Positives = 75/130 (57%)
Frame = +2
Query: 909 TEIPTLAVPDFSKVFVLETDASGTGLGAVLTQEGKPLAFWSATLSDRSQAKSVYERELMA 968
T P L +P+ +V+ TDAS TGLG VLTQ K +A+ S L ++ E+ A
Sbjct: 8 TSAPILVLPELI-TYVVYTDASITGLGCVLTQHEKVIAYASRQLRKHEGNYPTHDLEMAA 184
Query: 969 VVRAVQRWRHYLLGRHFIIRTDQKSLKFLTEQQVVGEGQFKWISKLSGYDFEIQYKPGKD 1028
VV A++ WR YL G I TD KSLK++ Q + Q +W+ ++ YD +I Y PGK
Sbjct: 185 VVFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDITYYPGKA 364
Query: 1029 NSAADAMSRR 1038
N ADA+SRR
Sbjct: 365 NLVADALSRR 394
>BF649369
Length = 631
Score = 85.9 bits (211), Expect = 1e-16
Identities = 46/168 (27%), Positives = 88/168 (52%)
Frame = +3
Query: 108 ENLQRTANAQTERWRKLDIPVFQGEEDAYSWIQKLERYFRLKGATEEEKVQAIMVALDGK 167
E ++ + N +K+ +P+F+G+ D +WI + E YF ++ ++ +V+ ++++G
Sbjct: 78 EYVESSQNESRLAGKKVKLPLFEGD-DPVAWITRAEIYFDVQNTPDDMRVKLSRLSMEGP 254
Query: 168 ALSWYQWWETCNQVTTWTKFKEAMLERFQLTSNSSPFAALLALKQEGSVEEFVGQFERFA 227
+ W+ + K K+A++ R+ +PF L L+Q GSVEEFV FE +
Sbjct: 255 TIHWFNLLMETEDDLSREKLKKALIARYDGRRLENPFEELSTLRQIGSVEEFVEAFELLS 434
Query: 228 GMLKGIDEEHYMDIFVNGLKEEIAAEIKLYEPKSLTIMVKKALMVEQK 275
+ + EE Y+ F++GLK I ++ P + M++ A VE +
Sbjct: 435 SQVGRLPEEQYLGYFMSGLKAHIRRRVRTLNPTTRMQMMRIAKDVEDE 578
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 72.8 bits (177), Expect = 1e-12
Identities = 50/171 (29%), Positives = 82/171 (47%), Gaps = 2/171 (1%)
Frame = +2
Query: 1104 IPILCKEFHASLLGGHSGFFRTYRRLA-AVVYWEGMKSDIRDFVAECDTCQRNKFDNLSP 1162
IP + H S GH +T ++ A +W M D F+++CD CQR N+S
Sbjct: 95 IPGILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHSFISKCDPCQRQ--GNIS* 268
Query: 1163 AGLL-QPLPVPTQVWTDVSMDFIGGLPKSNGKDTILVVVDRLTKYAHFIPLRHPFTAPEV 1221
+ Q + +V+ +DF+G P S ILV VD ++K+ I A V
Sbjct: 269 RNEMPQNFILEVEVFDVWGIDFMGPFPSSYNNKYILVAVDYVSKWVEAIA-SPTNDATVV 445
Query: 1222 AALFLHEVVRLHGFPTTIVSDRDSLFLSSFWKELFRVSGTQLKFSSAYHPQ 1272
+F + G P ++SD S F++ +++L + +G + K ++AYHPQ
Sbjct: 446 VKMFKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHKVATAYHPQ 598
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 71.2 bits (173), Expect = 3e-12
Identities = 33/65 (50%), Positives = 44/65 (66%)
Frame = -1
Query: 658 RPSISPYSSPVILVRKKDGSWRFCVDYRALNKITVPNKFPIPVIDELLDEIGPARVFSKI 717
+PSISP + ++ VRKKDG +R C+DYR NK+T NK+P+P ID L D+I F I
Sbjct: 262 QPSISP*GAALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCYF*NI 83
Query: 718 DLKSG 722
DL+ G
Sbjct: 82 DLRLG 68
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 56.2 bits (134), Expect = 9e-08
Identities = 30/63 (47%), Positives = 38/63 (59%), Gaps = 1/63 (1%)
Frame = +2
Query: 1389 DEVFLKLQPYRRRSLAKKMNEKLSPRYYGPYPIVAKIGAVAYRLEL-PAHSRVHPVFHVS 1447
++V LK+ P R KLS RY GP+ ++ +IG VAY L L P S VHPVFHVS
Sbjct: 2 EQVLLKVLPTERGDCRFGKRGKLSLRYIGPFEVIKRIGEVAYELALPPGLSGVHPVFHVS 181
Query: 1448 LLK 1450
+ K
Sbjct: 182 MFK 190
>TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulchellus},
partial (7%)
Length = 2304
Score = 52.4 bits (124), Expect = 1e-06
Identities = 37/154 (24%), Positives = 71/154 (46%), Gaps = 21/154 (13%)
Frame = +2
Query: 63 VATMTRRRAGEHGDSDGEQERRRMEEERTVNGGQPGSVRDEADARE--------NLQRTA 114
+ M RR E + E RR++++ + + Q + E E + R++
Sbjct: 401 IVKMPPRRQNERSLQEMEDMRRQIQQLQEIINAQQALLEAEQRRFEGDVSYSDSSSSRSS 580
Query: 115 NAQTERWR----KLDIPVFQGE---EDAYSWIQKLERYFRLKGATEEEKVQAIMVALDGK 167
++Q + + K+DIP F+G +D W+Q +ER F K EE+KV+ + L
Sbjct: 581 HSQRRQLQMNDIKVDIPDFEGNLQLDDFLDWLQTIERVFEYKEVPEEQKVKIVAAKLKKH 760
Query: 168 ALSWYQ------WWETCNQVTTWTKFKEAMLERF 195
AL W++ E +++ TW K ++ + ++
Sbjct: 761 ALIWWENLKRRRKREGKSKIKTWDKMRQKLTRKY 862
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 50.1 bits (118), Expect = 7e-06
Identities = 33/123 (26%), Positives = 60/123 (47%), Gaps = 12/123 (9%)
Frame = +2
Query: 1408 NEKLSPRYYGPYPIVAKIGAVAYRLELPAH-SRVHPVFHVSLLKTAVGTNFQPQPLPAAL 1466
++KL+ R+ GPY I ++G VAYR+ LP H +H VFHVS L+ + P P
Sbjct: 23 SKKLTVRFIGPYQISERVGTVAYRVGLPPHLLNLHDVFHVSQLR-----KYVPDPSHVIQ 187
Query: 1467 NEDHE-----------LLVEPESVLAVRETTMGQIEVLIQWHSLPACENSWESAAQLHEA 1515
++D + + ++ V +R + + V+ W +WE +++ E+
Sbjct: 188 SDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVV--WDRANGESLTWELESKMVES 361
Query: 1516 FPD 1518
+P+
Sbjct: 362 YPE 370
>TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA POLYMERASE II
{Encephalitozoon cuniculi}, partial (0%)
Length = 1247
Score = 49.7 bits (117), Expect = 9e-06
Identities = 36/155 (23%), Positives = 73/155 (46%), Gaps = 21/155 (13%)
Frame = -2
Query: 62 IVATMTRRRAGEHGDSDGEQERRRMEEERTVNGGQPGSV-------RDEADARENLQ-RT 113
++ M RR E + E+ RR +++ + + Q + +D + ++L R+
Sbjct: 955 VIVQMPPRRQNERSLQEMEKMRR*IQQLQEIVNAQQALLEAQQKRFKDHVSSSDSLSSRS 776
Query: 114 ANAQTERWR----KLDIPVFQGE---EDAYSWIQKLERYFRLKGATEEEKVQAIMVALDG 166
+ +Q ++ K DIP F+G +D W+Q +ER F+ K EE+KV+ + L
Sbjct: 775 SRSQRREFQMNDIK*DIPDFEGNLQPDDLLDWLQIMERLFKYKEVLEEQKVKIVAAKLKK 596
Query: 167 KALSWYQ------WWETCNQVTTWTKFKEAMLERF 195
A W++ E +++ TW K ++ + ++
Sbjct: 595 LASIWWENVKRRRKREGKSKIKTWEKMRQKLTRKY 491
>BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent virus}, partial
(1%)
Length = 726
Score = 49.3 bits (116), Expect = 1e-05
Identities = 39/164 (23%), Positives = 79/164 (47%), Gaps = 10/164 (6%)
Frame = +2
Query: 42 RRERAIEHERVEQRFRNLEAIVATMTRRRAGEHGDSDGEQERRRMEEERTVNGGQPGSVR 101
+ +R + +E+ R ++ + T+ ++A E +RRR +++ GS
Sbjct: 137 QNQRPLHEIEMEEMRRQIQLLQETVNAQQALL------EAQRRRNDDD--------GSGS 274
Query: 102 DEADARENLQRTANAQTERWR-KLDIPVFQGE---EDAYSWIQKLERYFRLKGATEEEKV 157
D + +R + R+ QT + K+DIP F G+ ++ W+Q +ER F+ K EE+KV
Sbjct: 275 DSSSSRSS--RSHRRQTRMSKIKVDIPDF*GKLQPDEFVDWLQTIERVFKYKEVAEEQKV 448
Query: 158 QAIMVALDGKALSWY------QWWETCNQVTTWTKFKEAMLERF 195
+ + L A W+ + E +++ TW K ++ + ++
Sbjct: 449 KIVAAKLKKHASIWWKNLKRKRNCEGKSKIKTWDKMRQKLTRKY 580
>AA660473
Length = 655
Score = 43.5 bits (101), Expect = 6e-04
Identities = 54/187 (28%), Positives = 86/187 (45%), Gaps = 14/187 (7%)
Frame = -2
Query: 389 DGEEEEIETLEPLTE-EIEEAMELKHLSLNSIQGISSRKSLKVWGKLKGTAIVVLVDCGA 447
DGE+E+I + E+ E +LK + +L++ G LKG IV+LVD GA
Sbjct: 633 DGEKEDILVYNSMGLCEMTEFNKLK---------VVKPATLQLVGCLKGVPIVILVDIGA 481
Query: 448 THNFISQS-------LAREMELQITETPAYTVEVGDGHVVKSKGVCKNLSVQIQGFVV-T 499
HNF S R + Q +E ++ + G S+G ++GF
Sbjct: 480 NHNFDFASSVSDVGCKGRVILSQESEVG*WSENINVG*AY*SRGT-------VRGFHC*G 322
Query: 500 QDFFLFGLRGVDVVLGLEWLAGLGD-IKANFEELTLKFKLGNKKVVLKGEPELLK----K 554
D + L D+ LG+ WL LG+ + +++E T+ F+ + V L+G ++LK K
Sbjct: 321 VDAYELELGEFDMFLGVAWLEKLGN*VVFDWDERTICFEWKGEVVKLQG--QILKDWKIK 148
Query: 555 RASMNAL 561
R N L
Sbjct: 147 RGGKNEL 127
>BG580885
Length = 609
Score = 40.0 bits (92), Expect = 0.007
Identities = 39/171 (22%), Positives = 70/171 (40%), Gaps = 9/171 (5%)
Frame = +3
Query: 126 IPVFQG--EEDAYSWIQKLERYFRLKGATEEEKVQAIM-VALDGKALSWYQW-WETCNQV 181
+P+F+G E + + + + R A+ E + I V L+ ++ WY E
Sbjct: 45 LPIFRGTPNESPITHLSRFNKVCRANNASSVEMQKKIFPVTLEEESALWYDLNIEPYYIS 224
Query: 182 TTWTKFKEAMLERFQLTSNSSPFAALLALKQEGSVEEFVGQFERFAGMLK-----GIDEE 236
+W + K + L+ + + L +G E F R +LK G++++
Sbjct: 225 LSWDEIKLSFLQAYYEIEPVEELRSELMGIHQGEKERVRSYFLRLQWILKRWPEHGLEDD 404
Query: 237 HYMDIFVNGLKEEIAAEIKLYEPKSLTIMVKKALMVEQKNLAVSKAGIGST 287
+FVNGL+EE + + +P SL ++ E K I ST
Sbjct: 405 VIKGVFVNGLREEFHDWVLMQKPTSLNDALRLTFDFEYVRRISGKKEIVST 557
>TC77946 homologue to GP|10334491|emb|CAC10207. putative splicing factor
{Cicer arietinum}, complete
Length = 2061
Score = 38.5 bits (88), Expect = 0.020
Identities = 28/102 (27%), Positives = 41/102 (39%), Gaps = 1/102 (0%)
Frame = +3
Query: 23 DREEERRNREEDR-RIRDEERRERAIEHERVEQRFRNLEAIVATMTRRRAGEHGDSDGEQ 81
D +++R+ DR R RD E RER+ +G H + DGE+
Sbjct: 285 DENGSKKDRDRDRDRERDREHRERS------------------------SGRHRERDGER 392
Query: 82 ERRRMEEERTVNGGQPGSVRDEADARENLQRTANAQTERWRK 123
R E E+ V G RD +E +R + ER R+
Sbjct: 393 GSREKEREKDVERGGRDKERDREREKERERRDKEREKERERR 518
Score = 38.1 bits (87), Expect = 0.027
Identities = 33/117 (28%), Positives = 53/117 (45%), Gaps = 13/117 (11%)
Frame = +3
Query: 21 NRDREEERRNREEDR-------RIRDEERRERAIEHER-VEQRFRNLEAIVATMTRRRAG 72
+RDR+ +R E R R RD ER R E E+ VE+ R+ E R +
Sbjct: 306 DRDRDRDRERDREHRERSSGRHRERDGERGSREKEREKDVERGGRDKE---RDREREKER 476
Query: 73 EHGDSDGEQERRRMEEERTVNGG-----QPGSVRDEADARENLQRTANAQTERWRKL 124
E D + E+ER R ++ER ++E ++ QR A + +RW+++
Sbjct: 477 ERRDKEREKERERRDKEREKERA*ERKRGERETKEEYESS*ERQRRARFRKQRWQEI 647
Score = 37.0 bits (84), Expect = 0.060
Identities = 28/88 (31%), Positives = 41/88 (45%), Gaps = 1/88 (1%)
Frame = +3
Query: 4 SRMEARVEAIERFIEVMNRDREEER-RNREEDRRIRDEERRERAIEHERVEQRFRNLEAI 62
SR + R + +ER RDRE E+ R R + R ++ ERR++ E ER +R R
Sbjct: 396 SREKEREKDVERGGRDKERDREREKERERRDKEREKERERRDKEREKERA*ERKRG---- 563
Query: 63 VATMTRRRAGEHGDSDGEQERRRMEEER 90
R E+ S Q R R ++R
Sbjct: 564 ----ERETKEEYESS*ERQRRARFRKQR 635
>CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product {Drosophila
melanogaster}, partial (20%)
Length = 192
Score = 38.1 bits (87), Expect = 0.027
Identities = 21/58 (36%), Positives = 35/58 (60%), Gaps = 5/58 (8%)
Frame = +1
Query: 933 GLGAVLTQEGK-----PLAFWSATLSDRSQAKSVYERELMAVVRAVQRWRHYLLGRHF 985
G+GA L+Q+ + P+A+ S L+ + +V ERE +A + A++ +RHYL G F
Sbjct: 13 GIGAGLSQKDEENHEHPIAYASRLLTAAERNYTVVERECLAAIWAIRNFRHYLHGPKF 186
>TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Cicer
arietinum}, partial (8%)
Length = 516
Score = 37.7 bits (86), Expect = 0.035
Identities = 33/110 (30%), Positives = 49/110 (44%), Gaps = 9/110 (8%)
Frame = +2
Query: 127 PVFQGE---EDAYSWIQKLERYFRLKGATEEEKVQAIMVALDGKALSWY----QWWETCN 179
P F+G + A W++++ER FR+ E +KVQ L +A W+ E +
Sbjct: 134 PTFKGRYAPDGA*KWLKEIERIFRVMQCFETQKVQFGTHMLAEEADDWWISLLPVLEQDD 313
Query: 180 QVTTWTKFKEAMLER-FQLTSNSSPFAALLALKQ-EGSVEEFVGQFERFA 227
V TW F++ L R F L LKQ + SV E+ +F A
Sbjct: 314 AVVTWAMFRKEFLGRYFPEDVRGKKEIEFLELKQGDMSVTEYAAKFVELA 463
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 686
Score = 37.4 bits (85), Expect = 0.046
Identities = 25/91 (27%), Positives = 43/91 (46%)
Frame = -2
Query: 1233 HGFPTTIVSDRDSLFLSSFWKELFRVSGTQLKFSSAYHPQTDGQTEVVNRCLEVYLRCLT 1292
HG P IV+D S F+S+ ++E +L +S +PQ++GQ E N+ + L+
Sbjct: 685 HGLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKIIIDGLKKRL 506
Query: 1293 GSRPRKWVTCLPWAEFWFNSNYNRSAKMSPF 1323
+ W L + +RS K++ F
Sbjct: 505 DLKKGCWADELDGCSLVT*NKSSRSNKINSF 413
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,929,574
Number of Sequences: 36976
Number of extensions: 602264
Number of successful extensions: 3224
Number of sequences better than 10.0: 80
Number of HSP's better than 10.0 without gapping: 3130
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3202
length of query: 1553
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1444
effective length of database: 4,984,343
effective search space: 7197391292
effective search space used: 7197391292
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0318.12


