

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
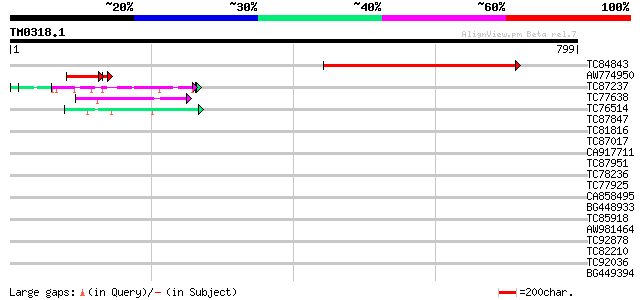
Query= TM0318.1
(799 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC84843 similar to GP|4567304|gb|AAD23715.1| unknown protein {Ar... 463 e-130
AW774950 72 8e-14
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 62 1e-09
TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protei... 47 3e-05
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 44 2e-04
TC87847 similar to GP|15929245|gb|AAH15068.1 Similar to adaptor-... 42 0.001
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 40 0.005
TC87017 similar to PIR|A96564|A96564 unknown protein 23094-2177... 40 0.005
CA917711 39 0.008
TC87951 similar to PIR|C86418|C86418 hypothetical protein AAG126... 38 0.017
TC78236 weakly similar to GP|15451010|gb|AAK96776.1 Unknown prot... 36 0.051
TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1 ... 35 0.15
CA858495 35 0.15
BG448933 similar to PIR|T09648|T09 nucleolin homolog nuM1 - alfa... 34 0.19
TC85918 similar to GP|13562014|gb|AAK30610.1 fibroin 1 {Plectreu... 34 0.19
AW981464 similar to GP|18376589|em putative cyclosporin A-bindin... 33 0.33
TC92878 homologue to PIR|F85062|F85062 hypothetical protein AT4g... 33 0.33
TC82210 weakly similar to GP|15450527|gb|AAK96556.1 AT5g21160/T1... 33 0.43
TC92036 homologue to EGAD|58716|61404 groucho-related gene 4 pro... 33 0.43
BG449394 similar to PIR|A83006|A830 hypothetical protein PA5121 ... 33 0.43
>TC84843 similar to GP|4567304|gb|AAD23715.1| unknown protein {Arabidopsis
thaliana}, partial (30%)
Length = 909
Score = 463 bits (1191), Expect = e-130
Identities = 225/278 (80%), Positives = 248/278 (88%)
Frame = +3
Query: 443 SRLVESVWWQALTPYMQSPAGDFSSNKSFGRVLGPALGDHNQGNFSINLWRYAFEDAFQR 502
++L+ + + ALTPYMQS GD SNKS GR+LGPALGDHNQGNFSINLWR AF+DAFQR
Sbjct: 3 NKLISFLCF*ALTPYMQSSVGDSCSNKSAGRLLGPALGDHNQGNFSINLWRNAFQDAFQR 182
Query: 503 LCPLRAGGHECGCLPVLARMVMEQCIARLDVAMFNAILRESALEIPTDPISDPILDSKVL 562
LCPLRAGGHECGCLPV+ARMVMEQCI RLDVAMFNAILRESALEIPTDPISDPI+DSKVL
Sbjct: 183 LCPLRAGGHECGCLPVMARMVMEQCIDRLDVAMFNAILRESALEIPTDPISDPIVDSKVL 362
Query: 563 PIPAGDLSFGSGAQLKNSVGNWSRWLTDMFGMDVEDCVQEYQDSGENDERQGGDGEPKPF 622
PIPAG+LSFGSGAQLKNSVGNWSR LTDMFG+D EDC +EY ++ ENDER+GG GE K F
Sbjct: 363 PIPAGNLSFGSGAQLKNSVGNWSRLLTDMFGIDAEDCSEEYPENSENDERRGGPGEQKSF 542
Query: 623 VLLNDLSDLLMLPKDMLIDRHVREEVCPSITLSLIIRVLCNFTPDEFCPDPVPGTVLEAL 682
LLNDLSDLLMLPKDML+DR V +EVCPSI+LSLIIRVLCNFTPDEFCPD VPG VLEAL
Sbjct: 543 ALLNDLSDLLMLPKDMLMDRQVSQEVCPSISLSLIIRVLCNFTPDEFCPDAVPGAVLEAL 722
Query: 683 NAETIAERRLSAESVRSFPYAAAAVVYVPPSSSNVAEK 720
N ETI ERR+SAES+RSFPY+AA VVY+PPSS NV +
Sbjct: 723 NGETIVERRMSAESIRSFPYSAAPVVYMPPSSVNVGRE 836
Score = 36.2 bits (82), Expect = 0.051
Identities = 24/56 (42%), Positives = 29/56 (50%), Gaps = 5/56 (8%)
Frame = +1
Query: 690 RRLSAESVRSFPYAAAAVVYVP-----PSSSNVAEKVAEAGGKSHLARNVSAVQRR 740
R L +E + P A+ + + P S AEKVAE K HL RNVSAV RR
Sbjct: 730 RLLWSEECQRSPLEASLIARLQLCICLPRQSMWAEKVAEGRRKCHLTRNVSAVHRR 897
>AW774950
Length = 550
Score = 72.0 bits (175), Expect(2) = 8e-14
Identities = 34/52 (65%), Positives = 43/52 (82%)
Frame = +3
Query: 81 VNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTVKDSVSSQGDSF 132
++RSEEAL +MKVN ++ N ++D EKEQKEGN +V DTDT KDS+SSQGD F
Sbjct: 351 LSRSEEALVKMKVNEVIDNHSTDLEKEQKEGNEDVYDTDTTKDSLSSQGDFF 506
Score = 23.5 bits (49), Expect(2) = 8e-14
Identities = 11/15 (73%), Positives = 12/15 (79%)
Frame = +1
Query: 131 SFTNEDEKVEKASKD 145
SFTNED VEKA K+
Sbjct: 502 SFTNEDV*VEKALKE 546
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 61.6 bits (148), Expect = 1e-09
Identities = 65/291 (22%), Positives = 115/291 (39%), Gaps = 23/291 (7%)
Frame = +1
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSN 61
K+ EK K G + + E+ ++ S + E E K+ Q+ N G+
Sbjct: 142 KDEEKSKQENEEIKDGEKIQQENEENKDEEKSQQENEENKDEEKSQQENELKKNEGGEKE 321
Query: 62 TAS--------ENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNG 113
T EN ET E D ++ + + A+ +V N D+++ +E NG
Sbjct: 322 TGEITEEKSKQENEETSETNSKDKENEESNQNGSDAKEQVGE---NHEQDSKQGTEETNG 492
Query: 114 ----EVSDTDTVKDSVSSQG---DSFTNEDEKVEKASKDAKSKVKVS--PSESSRGLKER 164
E + D +K+ SS D N + + E S D S V ESS +E+
Sbjct: 493 TEGGEKEEHDKIKEDTSSDNQVQDGEKNNEAREENYSGDNASSAVVDNKSQESSNKTEEQ 672
Query: 165 SDRKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKA------PVKASSESS 218
D+K +++ +QK +T+ +N+ N S+ +A P ++SES
Sbjct: 673 FDKKE---KNEFELESQKNSNETTESTDSTITQNSQGNESEKDQAQTENDTPKGSASESD 843
Query: 219 EGVDENHVLEVKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAAL 269
E E + ++ +Q+ E+ + + N + ED +AL
Sbjct: 844 EQKQEQEQNNTTKDDV-QTTDTSSQNGNDTTEKQNETSEDANSKKEDSSAL 993
Score = 55.5 bits (132), Expect = 8e-08
Identities = 64/291 (21%), Positives = 119/291 (39%), Gaps = 33/291 (11%)
Frame = +1
Query: 13 SQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGD---SNTASENSET 69
S G+ TE++ + NS K + + TP + + + + GD S T + +SET
Sbjct: 907 SSQNGNDTTEKQNETSEDANSKK--EDSSALNTTPNNEDSKSGVAGDQADSTTTTSSSET 1080
Query: 70 YENVVI--DYVDDVNRSEEALA------------------EMKVNAMVANQASDTEKEQK 109
+ +Y D N + E + + N + SDT EQK
Sbjct: 1081 QDGNTNHGEYKDTTNENPEKNSGQEGTQESGSSSNTFDNKDAASNKVQLTTTSDTSSEQK 1260
Query: 110 --EGNGEVSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDR 167
E + S +++ ++ ++ G S T DE +KD+ S+V S S+ G +
Sbjct: 1261 KDESSSAESKSESSQNDNANSGQSNTTSDESAND-NKDS-SQVTTSSENSAEGNSNTENN 1434
Query: 168 KTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVL 227
++ N N++ + N+N ++ +K + A +ES E EN+
Sbjct: 1435 SDENQNDSKNNENTNDSGNTSNDANVNENQNENAAQTKTSENEGDAQNESVESKKENNES 1614
Query: 228 EVKEIEILDGASNGAQSLGS--EDERHETVN----AEENGE--HEDKAALE 270
K+++ +++ S S +D++ V+ E NGE H D ++ E
Sbjct: 1615 AHKDVDNNSNSNDQGSSDTSVTQDDKESRVDLGTLPESNGESHHNDVSSAE 1767
Score = 45.4 bits (106), Expect = 8e-05
Identities = 43/208 (20%), Positives = 89/208 (42%), Gaps = 2/208 (0%)
Frame = +1
Query: 59 DSNTASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDT 118
+S E+SE ++ + + S E + ++ N+ + E++ K+ N E+ D
Sbjct: 28 ESKETGESSEEKSHLENEESKETGESSEEKSHLE------NEENKDEEKSKQENEEIKDG 189
Query: 119 DTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSD 178
+ + Q ++ N+DE + S E+ E ++ N+L K ++
Sbjct: 190 EKI------QQENEENKDE-------------EKSQQENEENKDEEKSQQENEL--KKNE 306
Query: 179 SNQKKPMNSTKGPSRVTNKNTSSNNSKPV--KAPVKASSESSEGVDENHVLEVKEIEILD 236
+K+ T+ S+ N+ TS NSK + + S++ E V ENH + K+
Sbjct: 307 GGEKETGEITEEKSKQENEETSETNSKDKENEESNQNGSDAKEQVGENHEQDSKQ----- 471
Query: 237 GASNGAQSLGSEDERHETVNAEENGEHE 264
G + G E E H+ + + + +++
Sbjct: 472 GTEETNGTEGGEKEEHDKIKEDTSSDNQ 555
>TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protein {Oryza
sativa (japonica cultivar-group)}, partial (62%)
Length = 2345
Score = 47.0 bits (110), Expect = 3e-05
Identities = 39/174 (22%), Positives = 74/174 (42%), Gaps = 11/174 (6%)
Frame = +1
Query: 94 NAMVANQASDTEKEQKEGNGEVSDTDTVK-----------DSVSSQGDSFTNEDEKVEKA 142
N V ++ KEQ E +VS+TD +++GDS + +EK E+
Sbjct: 439 NEDVPQESKSEVKEQTEVREQVSETDNSNARQFEDNPGDLPEDATKGDSNVSSEEKSEEN 618
Query: 143 SKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSSN 202
S + S+ + E + E S+ + NK + S + + K S NK+ S
Sbjct: 619 STEKSSEDTKTEDEGKKTEDEGSNTENNKDGEEASTKESESDESEKKDESEENNKSDSDE 798
Query: 203 NSKPVKAPVKASSESSEGVDENHVLEVKEIEILDGASNGAQSLGSEDERHETVN 256
+ K SS+S+E D N +V++ + + N ++ ++ + ++ N
Sbjct: 799 SE-------KKSSDSNETTDSNVEEKVEQSQNKESDENASEKNTDDNAKDQSSN 939
Score = 40.4 bits (93), Expect = 0.003
Identities = 43/201 (21%), Positives = 89/201 (43%), Gaps = 7/201 (3%)
Frame = +1
Query: 11 RNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNTASENSETY 70
+ S+++ +TE RE DNS+ E P+D + ++ V + ENS
Sbjct: 454 QESKSEVKEQTEVREQVSETDNSNARQFEDNP-GDLPEDATKGDSNVSSEEKSEENSTEK 630
Query: 71 ENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTE--KEQKEGNGEVSDTDTVKDSVSSQ 128
+ D+ ++E+ + S+TE K+ +E + + S++D + S+
Sbjct: 631 SSEDTKTEDEGKKTED-------------EGSNTENNKDGEEASTKESESDESEKKDESE 771
Query: 129 GDSFTNEDEKVEKAS-----KDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQKK 183
++ ++ DE +K+S D+ + KV S++ + S++ T+ +K SN+
Sbjct: 772 ENNKSDSDESEKKSSDSNETTDSNVEEKVEQSQNKESDENASEKNTDD-NAKDQSSNEVF 948
Query: 184 PMNSTKGPSRVTNKNTSSNNS 204
P + S + N+ T+ S
Sbjct: 949 PSGA---QSELLNETTTQTGS 1002
Score = 37.0 bits (84), Expect = 0.030
Identities = 38/152 (25%), Positives = 64/152 (42%), Gaps = 31/152 (20%)
Frame = +1
Query: 85 EEALAEMKVNAMVANQASDTE----KEQKEGNGEVSDTDTVKDS-VSSQGDSFTNEDEKV 139
+E+ +E+K V Q S+T+ ++ ++ G++ + T DS VSS+ S E+
Sbjct: 454 QESKSEVKEQTEVREQVSETDNSNARQFEDNPGDLPEDATKGDSNVSSEEKS---EENST 624
Query: 140 EKASKDAKSK---------------------VKVSPSESSRGLKERSDRKTNKLQS---- 174
EK+S+D K++ SES K+ + NK S
Sbjct: 625 EKSSEDTKTEDEGKKTEDEGSNTENNKDGEEASTKESESDESEKKDESEENNKSDSDESE 804
Query: 175 -KVSDSNQKKPMNSTKGPSRVTNKNTSSNNSK 205
K SDSN+ N + + NK + N S+
Sbjct: 805 KKSSDSNETTDSNVEEKVEQSQNKESDENASE 900
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 44.3 bits (103), Expect = 2e-04
Identities = 50/231 (21%), Positives = 88/231 (37%), Gaps = 36/231 (15%)
Frame = +3
Query: 78 VDDVNRSEEALAEMKVNAMVANQASDTEKEQ--------------KEGNGEVSDTDTVKD 123
V++V ++AL +K + ++S++E EQ K+GN + + +T
Sbjct: 237 VEEVAAKQKALKVVKKEESSSEESSESEDEQPVVKAPAPSKKTPAKKGNVKKAQPETT-- 410
Query: 124 SVSSQGDSFTNEDEKVEK------ASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVS 177
S S DS ++++E+V+K SK+ + K + +E SD V
Sbjct: 411 SEESDSDSSSSDEEEVKKPVSKAVPSKNGSAPAKKVDTSEEEDSEESSDEDKKPAAKAVP 590
Query: 178 DSNQKKPMNSTKGPSRVTNKNT---------------SSNNSKPV-KAPVKASSESSEGV 221
N P T++++ S N S P KA ++S E SE
Sbjct: 591 SKNGSAPAKKAASDEEDTDESSDEDEEDEKPAAKAVPSKNGSVPAKKADTESSDEDSESS 770
Query: 222 DENHVLEVKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELK 272
DE + A + S++E E + +E+ + K A K
Sbjct: 771 DEEDKKPAAKASKNVSAPTKKAASSSDEESDEESDEDEDAKPVSKPAAVAK 923
>TC87847 similar to GP|15929245|gb|AAH15068.1 Similar to adaptor-related
protein complex AP-3 beta 1 subunit {Mus musculus},
partial (2%)
Length = 931
Score = 41.6 bits (96), Expect = 0.001
Identities = 49/219 (22%), Positives = 80/219 (36%), Gaps = 8/219 (3%)
Frame = +1
Query: 56 LVGDSNTASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEV 115
LV D TAS + E + + + +V Q D+ E++EG+ E
Sbjct: 88 LVDDPPTASSSDE---------------EQPPSTQKPADKVVNEQDEDSSSEEEEGDEED 222
Query: 116 SDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVS---PSESSRGLKERSDRKTNKL 172
+ ++ +Q ++D K ASK+ +S P+ES G + S+ T
Sbjct: 223 GSSSEDEEEEDNQTQQTPSKDSK--PASKNPPPSTPISNPKPAESESGSESGSESGTESD 396
Query: 173 QSKVSDSNQKKPMNS-----TKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVL 227
S P N P + + + + P+K+ K +ESS G D
Sbjct: 397 SEPEHKSEPTPPPNPKVKPLASKPMKTQTQAQAQSTPVPIKSGTKRVAESSTGNDSKR-- 570
Query: 228 EVKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDK 266
K+ G GS+DE +A+ GE K
Sbjct: 571 SKKKTTTAGG--------GSDDENEVEEDAKLTGEDSKK 663
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 39.7 bits (91), Expect = 0.005
Identities = 34/183 (18%), Positives = 71/183 (38%), Gaps = 5/183 (2%)
Frame = +3
Query: 5 EKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNTAS 64
+K + ++ + K + + +++K ++ KT ++G + K + + + N
Sbjct: 126 DKVEIEKDLEIKSVEKDDEKKEKKDKEKKDKTDVDEGKDKKDKEKKKKEKK---EENVKG 296
Query: 65 ENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTVKDS 124
E + D+ E+ E K EK+ K+ + D + D
Sbjct: 297 EEEDG---------DEKKDKEKKKKEKKEKGKEDKDKDGEEKKSKKDKEKKKDKNEDDDE 449
Query: 125 VSSQGDSFTNEDEKVEKASKDAKSKVKVSP-----SESSRGLKERSDRKTNKLQSKVSDS 179
N+D+K +K +D K + KVS E+++ KE+ +K +K + K
Sbjct: 450 GEDGSKKKKNKDKKEKKKEEDEKEEGKVSVRDIDIEETAKEGKEKKKKKEDKEEKKKKSG 629
Query: 180 NQK 182
K
Sbjct: 630 KDK 638
>TC87017 similar to PIR|A96564|A96564 unknown protein 23094-21772
[imported] - Arabidopsis thaliana, partial (30%)
Length = 1701
Score = 39.7 bits (91), Expect = 0.005
Identities = 49/226 (21%), Positives = 93/226 (40%), Gaps = 24/226 (10%)
Frame = +3
Query: 3 ETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTP------QDRSAANNL 56
++ K++A+ T+ + + +D + + K +E+ ++ Q +SA N
Sbjct: 135 QSSKKRAAGRELTRDTPIDDEEDDTDFEAGTFKKASEEVLATRRMVKVVRRQQKSAPNPF 314
Query: 57 VG------DSNTASENSETYENVVIDYVDDVNRSEEALA-EMKVNAMVANQASDTEKEQK 109
G +TA T E D+ + E A E++ + N +D K
Sbjct: 315 AGIRLPAPTESTAKSGEATSETQPEKAKDEETKQPEIKAPEVEDKSTSNNDVADRNNAGK 494
Query: 110 EGNGEVSDTDTVK--DSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDR 167
+ + +D+D K + S G NED+K E A K++ ++ P + E SD+
Sbjct: 495 DLAEKETDSDNSKVDNEQSKDGSKIENEDKK-EVADKESAGEINKEPPTEEKN-TENSDK 668
Query: 168 KTNK----LQSKVSDSNQKKPMNSTK-----GPSRVTNKNTSSNNS 204
N + KVS+S K+ S + GP + + +S+ N+
Sbjct: 669 NGNSESKDKEDKVSESKDKEDKVSDEPTAEGGPFKSFQQLSSNQNA 806
>CA917711
Length = 516
Score = 38.9 bits (89), Expect = 0.008
Identities = 44/174 (25%), Positives = 75/174 (42%), Gaps = 4/174 (2%)
Frame = +1
Query: 10 SRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAA-NNLVGDSNTASENSE 68
S + T GSR ++ + +N+K + K +S++ N G T ENS
Sbjct: 10 SLSGLTSGSRNNYEHNEETRNSGQNCIVNDKRVQQKINMSKSSSFNKFSGVGRTGKENSH 189
Query: 69 TYENVVIDYVDDVNRSEEALAEM-KVNAMVANQASDTEKEQKEGNGEVSDTDTVKDSVSS 127
+ V V N SE ++ KVN ++ S TEK+ D +K+ +S
Sbjct: 190 S----VWQKVQKNNSSECGGGDLKKVNTTLSQSVSATEKD---------DPSAIKNCNNS 330
Query: 128 QGDSFTN--EDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDS 179
G + + ED+K K KSK K + S +G S + +N ++ ++D+
Sbjct: 331 VGANAVSGPEDKKNVKNKVSRKSKGK-TDSVPRKGACNYSRKGSNFNRTVLNDN 489
>TC87951 similar to PIR|C86418|C86418 hypothetical protein AAG12629.1
[imported] - Arabidopsis thaliana, partial (13%)
Length = 922
Score = 37.7 bits (86), Expect = 0.017
Identities = 47/198 (23%), Positives = 78/198 (38%), Gaps = 3/198 (1%)
Frame = +3
Query: 23 RREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNTASENSETYENVVIDYVDDVN 82
+ E+ L + +S K LN ++ +N+++ + E+Y N V + DD+
Sbjct: 378 KSEESLEKIDSPKDLNYGSRKNN-----GGSNDVIETGTAKNSKDESYNNSVAEVSDDLV 542
Query: 83 RSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTVKDSVSSQGDSFTNEDEKVEKA 142
++ ++ EM+ TV D + S E + K
Sbjct: 543 KAVTSVTEMQSGG------------------------TVNDILEKPLGSRAAETDLAVKR 650
Query: 143 SKDAKSKVKVSPSESSR--GLKERSDRK-TNKLQSKVSDSNQKKPMNSTKGPSRVTNKNT 199
++D KV V P E R LKE R+ K+ S VS S+ + N + + +N
Sbjct: 651 NED---KVHVLPDEGGRISSLKEPETREFDGKVSSSVSQSSIPEATNDVEN-VKGSNAAE 818
Query: 200 SSNNSKPVKAPVKASSES 217
SS N PV AP+ S
Sbjct: 819 SSENQPPV-APLMVHKTS 869
>TC78236 weakly similar to GP|15451010|gb|AAK96776.1 Unknown protein
{Arabidopsis thaliana}, partial (61%)
Length = 1303
Score = 36.2 bits (82), Expect = 0.051
Identities = 45/153 (29%), Positives = 71/153 (45%), Gaps = 16/153 (10%)
Frame = +1
Query: 172 LQSKVSDSNQKKPM------NST---KGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVD 222
L S V +SN K M +ST GPS+ + +++SK + V+ + ++
Sbjct: 229 LDSTVMESNPSKRMEMEYSLDSTVLENGPSKRLRTESYASSSKAGREKVRRDKLNDRFME 408
Query: 223 ENHVLE------VKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEM 276
+ VLE ++ +L+ A L +E ER + E N E +K ELK E+
Sbjct: 409 LSSVLEPDTLPKTDKVSLLNDAVRVVTQLRNEAERLK----ERNDELREKVK-ELKAEKK 573
Query: 277 ELRVEKLEEEL-REVAALEVSLYSVVPEHGSSA 308
ELR EK + +L +E +V L SV S+A
Sbjct: 574 ELRDEKNKLKLDKEKLEQQVKLASVQSNFLSNA 672
>TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1
{Arabidopsis thaliana}, partial (47%)
Length = 1538
Score = 34.7 bits (78), Expect = 0.15
Identities = 48/189 (25%), Positives = 75/189 (39%), Gaps = 30/189 (15%)
Frame = +3
Query: 114 EVSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKV----SPSESSRGLKERSDRKT 169
+V +K S+ +S +++ E K KS K S SES +++R RK+
Sbjct: 507 DVKKDKEIKQKAKSESESEESKESDSELRKKRRKSYKKSRESDSESESESEVEDRKRRKS 686
Query: 170 NKLQSKVSDSN----------QKKPMNSTKGPSRVTNKNTSSNNS--------------- 204
K SD+N ++K + SR + + S+ S
Sbjct: 687 RKYSESDSDTNSEEEDRKRRRRRKSSRRRRKSSRKKRRCSDSDESETDDESGYDDSGRKR 866
Query: 205 KPVKAPVKASSESSEGVDENHVLEVKEIEILDGASNGAQSLGSE-DERHETVNAEENGEH 263
K K K+ +SSE V E +EIE+ G+ + + E D+ E AE N E
Sbjct: 867 KRSKRSRKSKKKSSEPVSEGS----EEIEL--GSDSAVAKINEEIDDVDEEKMAEMNAE- 1025
Query: 264 EDKAALELK 272
AL+LK
Sbjct: 1026----ALKLK 1040
>CA858495
Length = 763
Score = 34.7 bits (78), Expect = 0.15
Identities = 52/246 (21%), Positives = 104/246 (42%), Gaps = 18/246 (7%)
Frame = +3
Query: 54 NNLVGDSNTASENSETYENVVIDYVDDVNR--SEEALAEMKVNAMVANQASDTEKEQ--- 108
N+L+ +E + N V+ VN +E+ E KVN + +N +S +EKE
Sbjct: 30 NSLLQKEVGLAEKTNLLLNEKAGLVEKVNMLLNEKEGLEQKVNILESNLSSFSEKEAGSV 209
Query: 109 -------KEGNGEVSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGL 161
KE G + ++ ++SS + T + ++ + + S + + G
Sbjct: 210 DTTNLLLKEKEGLEQKLNILESNLSSFSEKETGLEMRIAQLQSETNSLL-----QKETGF 374
Query: 162 KERSDRKTNK--LQSKVSDSNQKKPMNSTKGPSRVTNKNTSS----NNSKPVKAPVKASS 215
E++++ N+ + S DS ++K +++ S K S+ +N + ++
Sbjct: 375 VEKTNQLLNEKNILSLKVDSLERKIIHAESDLSSFVEKENSTEEVISNLNGSISMLQGQV 554
Query: 216 ESSEGVDENHVLEVKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEE 275
E N +LE +++ +S+ +Q ++D A EN E L+ +IEE
Sbjct: 555 AELEESKNNLLLENQQLRENSSSSSASQDASAKD------RASENEE------LKSQIEE 698
Query: 276 MELRVE 281
+ VE
Sbjct: 699 AYMLVE 716
>BG448933 similar to PIR|T09648|T09 nucleolin homolog nuM1 - alfalfa, partial
(29%)
Length = 663
Score = 34.3 bits (77), Expect = 0.19
Identities = 38/162 (23%), Positives = 68/162 (41%), Gaps = 15/162 (9%)
Frame = +2
Query: 78 VDDVNRSEEALAEMKVNAMVANQASDTEKEQ--------------KEGNGEVSDTDTVKD 123
V++V ++AL +K + ++S++E EQ K+GN + + +T
Sbjct: 215 VEEVAAKQKALKVVKKEESSSEESSESEDEQPVVKAPAPSKKTPAKKGNVKKAQPETT-- 388
Query: 124 SVSSQGDSFTNEDEKVEK-ASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQK 182
S S DS ++++E+V+K SK SK +P++ KV S++
Sbjct: 389 SEESDSDSSSSDEEEVKKPVSKAVPSKNGSAPAK------------------KVDTSDED 514
Query: 183 KPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDEN 224
K + PS+ N S P K ++ E DE+
Sbjct: 515 KKPAAKAVPSK--------NGSAPAKKAASDEEDTDESSDED 616
>TC85918 similar to GP|13562014|gb|AAK30610.1 fibroin 1 {Plectreurys tristis},
partial (8%)
Length = 4117
Score = 34.3 bits (77), Expect = 0.19
Identities = 56/294 (19%), Positives = 111/294 (37%), Gaps = 4/294 (1%)
Frame = -1
Query: 1 MKETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDS 60
++ TE K + + T + E + +++T+ E+ ++ + V
Sbjct: 2095 VEATETVKDEPAATEVEATETVKEEPVATETEATETVKEEPVATEVEATETVKEESVATE 1916
Query: 61 NTASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDT 120
A+E + + ++ + E A E + V ++ TE V T+T
Sbjct: 1915 VEATETVKEEPVTTVVEPTEMVKDEPAATEFEATETVKDETIVTE---------VDPTET 1763
Query: 121 VKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSN 180
VK+ + T ++ D VKV P + +E + + + +++
Sbjct: 1762 VKEEAVATEVEITKTVKEPLVTEVDLTESVKVKPVVTEVDPREMVKEEPVATEVQATETV 1583
Query: 181 QKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILDGASN 240
+++P+ + P+ PV E ++ E H E +E E + +S
Sbjct: 1582 KEEPVATEVEPTETVKGE-----------PVVTEVEENQKEPEQHSTERREEEQPNASSI 1436
Query: 241 GAQSLGSEDERHETVNAEENG-EHEDKAAL--ELKIEEM-ELRVEKLEEELREV 290
QS E ++ + EE E E +AA+ E K +E EK+E + EV
Sbjct: 1435 PEQS----TETNDVIAVEEKSRELEFEAAILKETKNDEAGPAETEKVEPVVTEV 1286
>AW981464 similar to GP|18376589|em putative cyclosporin A-binding protein
{Picea abies}, complete
Length = 692
Score = 33.5 bits (75), Expect = 0.33
Identities = 24/78 (30%), Positives = 35/78 (44%)
Frame = +3
Query: 684 AETIAERRLSAESVRSFPYAAAAVVYVPPSSSNVAEKVAEAGGKSHLARNVSAVQRRGYT 743
A T + RLS S P +A PP S + + +A SHL +NV A+ G
Sbjct: 408 ASTSSSARLSRVLTSSRPSRRSAPAAAPPPSRSPSPSLASCRRLSHLNKNVHAI*DIGMN 587
Query: 744 SDEELEELDSPLTSIIDK 761
E +S T+++DK
Sbjct: 588 F*AAYENDESRHTTLLDK 641
>TC92878 homologue to PIR|F85062|F85062 hypothetical protein AT4g04980
[imported] - Arabidopsis thaliana, partial (1%)
Length = 608
Score = 33.5 bits (75), Expect = 0.33
Identities = 33/119 (27%), Positives = 56/119 (46%), Gaps = 11/119 (9%)
Frame = +2
Query: 118 TDTVKD-SVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRK--TNKLQS 174
TDT K + +++ +S ++ EK+E K K + SSRG K + D K T ++
Sbjct: 44 TDTCKIMNPNAEIESVSSNQEKLESKKKMKKVRSMKLQRLSSRGRKPQYDHKVRTTEMVE 223
Query: 175 KVSDSNQKKPMNS--TKGPSRVTNKNTSSNNS------KPVKAPVKASSESSEGVDENH 225
+ S + K +S G + + S+ S K K+ K SES++G+D N+
Sbjct: 224 EASPNYMKATGSSHAKDGFQIIQKRKMKSSRSIKLLTVKGPKSTTKLYSESTDGIDGNN 400
>TC82210 weakly similar to GP|15450527|gb|AAK96556.1 AT5g21160/T10F18_190
{Arabidopsis thaliana}, partial (24%)
Length = 1436
Score = 33.1 bits (74), Expect = 0.43
Identities = 27/99 (27%), Positives = 47/99 (47%), Gaps = 4/99 (4%)
Frame = +2
Query: 107 EQKEGNGEVSDTDTVKDS-VSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERS 165
+ GN S +D V S +S N D V+K + +++ + S +S+ + +S
Sbjct: 344 QTSSGNSGSSVAQVQQDQHVESTANSCQNSDTVVDKTKESSEATLNDSAHDSTSTEQNQS 523
Query: 166 DRKTNKLQSKVSDSNQKKPMNSTKGPS---RVTNKNTSS 201
++ T +VSD NQK+ NS + V NKN ++
Sbjct: 524 NKDT----FEVSDVNQKQDTNSHPSKNISHAVMNKNATT 628
>TC92036 homologue to EGAD|58716|61404 groucho-related gene 4 protein (Grg4)
{Mus musculus}, partial (3%)
Length = 1046
Score = 33.1 bits (74), Expect = 0.43
Identities = 47/212 (22%), Positives = 87/212 (40%), Gaps = 10/212 (4%)
Frame = -2
Query: 17 GSRRTERREDK--LHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNTASENSETYENVV 74
GS + E ++DK + ++N + + + +D + +N +T +SET E +
Sbjct: 1018 GSGQEELQDDKTGVSEENLDQPTETQMVSDEMQRDHTEIDN---QQSTLPLSSETEEGEM 848
Query: 75 IDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTVK----DSVSSQGD 130
+ D + + + NQ S E + V D D ++ +S D
Sbjct: 847 LPEAGDPEGGFDG-------SNMENQES-REATPEPSPARVDDDDALEAGEINSPEISTD 692
Query: 131 SFTNEDEKVEKASKDAKSKV----KVSPSESSRGLKERSDRKTNKLQSKVSDSNQKKPMN 186
+E + E A+ D K+ K + ES + ++ + LQS V++S+ K
Sbjct: 691 DKNDEGDLAEDAA-DGSDKLADVNKATSVESDQVVEPAPVASESNLQSSVAESSSSKLPV 515
Query: 187 STKGPSRVTNKNTSSNNSKPVKAPVKASSESS 218
S +G +R +K + PVK P S+ S
Sbjct: 514 SKQGATRSPSKTEDVKPTSPVK-PTSPISDMS 422
>BG449394 similar to PIR|A83006|A830 hypothetical protein PA5121 [imported] -
Pseudomonas aeruginosa (strain PAO1), partial (1%)
Length = 684
Score = 33.1 bits (74), Expect = 0.43
Identities = 40/194 (20%), Positives = 67/194 (33%), Gaps = 3/194 (1%)
Frame = +1
Query: 107 EQKEGNGEVSDTDTVKDSVSSQGDSFTNEDE--KVEKASKDAKSKVKVSPSESSRGLKER 164
E G G V+ D S D+ ++ +++K +K+A+ + K P +G
Sbjct: 130 ENSYGVGVVNRYALFLDDESDPLDALKAREQAKELKKKTKEAEKENKGKPETKPKGGAPA 309
Query: 165 SDRKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDEN 224
+ + + Q+ S N+ KGP+R +NT P + G EN
Sbjct: 310 TRKGIKETQNVKSQENRSGEQQKAKGPARPNERNTER------PPPRRREDRPQNGAVEN 471
Query: 225 HVLEVKEIEILDGASN-GAQSLGSEDERHETVNAEENGEHEDKAALELKIEEMELRVEKL 283
DGA + G +E N +N + ++ E E R
Sbjct: 472 K----------DGAPRPPRREFGDRRPNYERRNFSDNPDGAERRGPXPXREPREARDGPR 621
Query: 284 EEELREVAALEVSL 297
+R VSL
Sbjct: 622 RARVRTTIXASVSL 663
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.309 0.127 0.354
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,733,617
Number of Sequences: 36976
Number of extensions: 269384
Number of successful extensions: 1426
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 1397
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1421
length of query: 799
length of database: 9,014,727
effective HSP length: 104
effective length of query: 695
effective length of database: 5,169,223
effective search space: 3592609985
effective search space used: 3592609985
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0318.1


