

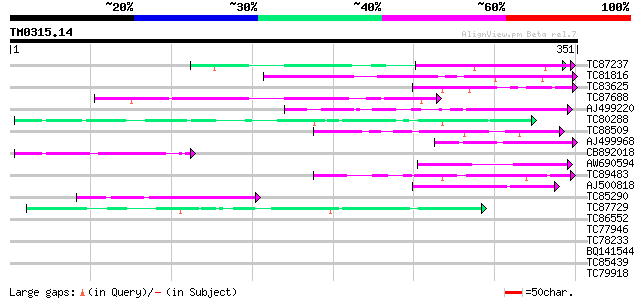
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0315.14
(351 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 60 1e-09
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 57 1e-08
TC83625 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 53 2e-07
TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~un... 50 1e-06
AJ499220 similar to SP|Q09863|YAF Hypothetical protein C29E6.10c... 47 8e-06
TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein... 46 2e-05
TC88509 similar to PIR|T01826|T01826 microfibril-associated prot... 45 3e-05
AJ499968 weakly similar to GP|23495961|gb| hypothetical protein ... 45 4e-05
CB892018 45 5e-05
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 43 2e-04
TC89483 similar to GP|18377662|gb|AAL66981.1 unknown protein {Ar... 43 2e-04
AJ500818 weakly similar to GP|10435304|dbj unnamed protein produ... 42 5e-04
TC85290 similar to GP|7767653|gb|AAF69150.1| F27F5.2 {Arabidopsi... 42 5e-04
TC87729 similar to GP|9759461|dbj|BAB10377.1 gene_id:MAF19.15~un... 41 6e-04
TC86552 similar to PIR|G85436|G85436 hypothetical protein AT4g36... 40 0.001
TC77946 homologue to GP|10334491|emb|CAC10207. putative splicing... 40 0.001
TC78233 similar to GP|15809838|gb|AAL06847.1 AT5g51120/MWD22_6 {... 40 0.001
BQ141544 similar to GP|7022293|dbj| unnamed protein product {Hom... 40 0.001
TC85439 homologue to SP|P36181|HS80_LYCES Heat shock cognate pro... 40 0.002
TC79918 similar to PIR|T06671|T06671 hypothetical protein T17F15... 37 0.009
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 60.5 bits (145), Expect = 1e-09
Identities = 35/100 (35%), Positives = 56/100 (56%), Gaps = 1/100 (1%)
Frame = +1
Query: 252 VEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKE-LKHGKELEMNISRELESGIRDGLD 310
+E EE +E GE +++ H E EE +E +EEK L++ + + S++ I+DG
Sbjct: 16 LENEESKETGESSEEKSHLENEESKETGESSEEKSHLENEENKDEEKSKQENEEIKDGEK 195
Query: 311 GDEEEEREGDEEKEVKHVKEKKKIEKLEKENEEKKVKRGK 350
+E E DEEK + +E K EK ++ENE KK + G+
Sbjct: 196 IQQENEENKDEEKSQQENEENKDEEKSQQENELKKNEGGE 315
Score = 47.8 bits (112), Expect = 6e-06
Identities = 55/242 (22%), Positives = 95/242 (38%), Gaps = 9/242 (3%)
Frame = +1
Query: 113 EEEREIGEVDEEK---EVKLGKKFGMAWIQVNHYELREHGERRKCGLLEFNFSIDLDSGI 169
EE +E GE EEK E + K+ G + + +H E E+
Sbjct: 25 EESKETGESSEEKSHLENEESKETGESSEEKSHLENEEN--------------------- 141
Query: 170 RDEIDEDGEEEREADGLHVDEENEREVDDGNEETELKYDKGYEMVRILVNHNVLKEQYMD 229
+DE E E DG + +ENE D+ + E + +K E K Q +
Sbjct: 142 KDEEKSKQENEEIKDGEKIQQENEENKDEEKSQQENEENKDEE-----------KSQQEN 288
Query: 230 KYKQEKRYNSKNLIVEYDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKE--L 287
+ K+ E E+E GE+ +++ E EE E + ++E E
Sbjct: 289 ELKK-------------------NEGGEKETGEITEEKSKQENEETSETNSKDKENEESN 411
Query: 288 KHGKELEMNISRELESGIRDGLDGDEEEEREGDEEKEVKHVKE----KKKIEKLEKENEE 343
++G + + + E + G +E EG E++E +KE +++ EK NE
Sbjct: 412 QNGSDAKEQVGENHEQDSKQGT--EETNGTEGGEKEEHDKIKEDTSSDNQVQDGEKNNEA 585
Query: 344 KK 345
++
Sbjct: 586 RE 591
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 56.6 bits (135), Expect = 1e-08
Identities = 53/210 (25%), Positives = 105/210 (49%), Gaps = 16/210 (7%)
Frame = +3
Query: 158 EFNFSIDLDSGIRDEIDEDGEE-EREADGLHVDEENER-EVDDGNEETELKYDKGYEMVR 215
E N I +D ++ ED E E++ + V++++E+ E D ++ + D+G +
Sbjct: 72 ESNIPIKIDDKSAGDVKEDKVEIEKDLEIKSVEKDDEKKEKKDKEKKDKTDVDEGKDK-- 245
Query: 216 ILVNHNVLKEQYMDKYKQEKRYNSKNLIVEYDGLDRVEEEEEREVGEVEKDRLHGEEEEE 275
DK K++K +N+ E + D +++E+++ + EK G+E+++
Sbjct: 246 ------------KDKEKKKKEKKEENVKGEEEDGDEKKDKEKKKKEKKEK----GKEDKD 377
Query: 276 REVSVVNEEKELKHGKELEMNISR---ELESGIRDGLDGDEEEEREGDEEKEVKHV---- 328
++ EEK+ K KE + + + E E G + + D++E+++ ++EKE V
Sbjct: 378 KD----GEEKKSKKDKEKKKDKNEDDDEGEDGSKKKKNKDKKEKKKEEDEKEEGKVSVRD 545
Query: 329 -------KEKKKIEKLEKENEEKKVKRGKD 351
KE K+ +K +++ EEKK K GKD
Sbjct: 546 IDIEETAKEGKEKKKKKEDKEEKKKKSGKD 635
Score = 35.0 bits (79), Expect = 0.043
Identities = 34/142 (23%), Positives = 66/142 (45%), Gaps = 14/142 (9%)
Frame = +3
Query: 4 EKRMKNENTRCKMKNEKPFSSDLESGLRDRRDGDGEEDREKVKVNEEKEVDHGKKLEMWN 63
+K +E K K +K E+ + DGD ++D+EK K ++++ K +
Sbjct: 213 DKTDVDEGKDKKDKEKKKKEKKEENVKGEEEDGDEKKDKEKKKKEKKEKGKEDKDKDGEE 392
Query: 64 KMDYKERKKIREKMVRKCRRAGLDG-------DGEKEREAGEDLESGRRDGLD------- 109
K K+++K ++K + G DG D +++++ ++ E G+ D
Sbjct: 393 KKSKKDKEKKKDK--NEDDDEGEDGSKKKKNKDKKEKKKEEDEKEEGKVSVRDIDIEETA 566
Query: 110 GYGEEEREIGEVDEEKEVKLGK 131
G+E+++ E EEK+ K GK
Sbjct: 567 KEGKEKKKKKEDKEEKKKKSGK 632
>TC83625 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (9%)
Length = 908
Score = 52.8 bits (125), Expect = 2e-07
Identities = 39/109 (35%), Positives = 65/109 (58%), Gaps = 7/109 (6%)
Frame = +2
Query: 250 DRVEEEEEREVGEVEKD--RLHGEEEEEREVSVVNE-----EKELKHGKELEMNISRELE 302
D+VE E++ E+ VEKD + +++E+++ + V+E +KE K ++ E N+ E E
Sbjct: 518 DKVEIEKDLEIKSVEKDDEKKEKKDKEKKDKTDVDEGKDKKDKEKKKKEKKEENVKGEEE 697
Query: 303 SGIRDGLDGDEEEEREGDEEKEVKHVKEKKKIEKLEKENEEKKVKRGKD 351
DGDE++ D+EK+ K KEK K +K +K+ EEKK K+ K+
Sbjct: 698 -------DGDEKK----DKEKKKKEKKEKGKEDK-DKDGEEKKSKKDKE 808
Score = 35.4 bits (80), Expect = 0.033
Identities = 33/149 (22%), Positives = 73/149 (48%), Gaps = 2/149 (1%)
Frame = +2
Query: 158 EFNFSIDLDSGIRDEIDEDGEE-EREADGLHVDEENER-EVDDGNEETELKYDKGYEMVR 215
E N I +D ++ ED E E++ + V++++E+ E D ++ + D+G +
Sbjct: 464 ESNIPIKIDDKSAGDVKEDKVEIEKDLEIKSVEKDDEKKEKKDKEKKDKTDVDEGKDK-- 637
Query: 216 ILVNHNVLKEQYMDKYKQEKRYNSKNLIVEYDGLDRVEEEEEREVGEVEKDRLHGEEEEE 275
DK K++K +N+ E + D +++E+++ + EK G+E+++
Sbjct: 638 ------------KDKEKKKKEKKEENVKGEEEDGDEKKDKEKKKKEKKEK----GKEDKD 769
Query: 276 REVSVVNEEKELKHGKELEMNISRELESG 304
++ EEK+ K KE + + + + + G
Sbjct: 770 KD----GEEKKSKKDKEKKKDKNEDDDGG 844
Score = 33.9 bits (76), Expect = 0.097
Identities = 49/194 (25%), Positives = 80/194 (40%)
Frame = +2
Query: 18 NEKPFSSDLESGLRDRRDGDGEEDREKVKVNEEKEVDHGKKLEMWNKMDYKERKKIREKM 77
+ K S++ + D+ GD +ED KV++ ++ E+ K +E K D K+ KK +EK
Sbjct: 449 SSKMEESNIPIKIDDKSAGDVKED--KVEIEKDLEI---KSVE---KDDEKKEKKDKEKK 604
Query: 78 VRKCRRAGLDGDGEKEREAGEDLESGRRDGLDGYGEEEREIGEVDEEKEVKLGKKFGMAW 137
+ G D +KE++ E E + GEEE + DE+K+ + KK
Sbjct: 605 DKTDVDEGKDKK-DKEKKKKEKKEENVK------GEEE----DGDEKKDKEKKKK----- 736
Query: 138 IQVNHYELREHGERRKCGLLEFNFSIDLDSGIRDEIDEDGEEEREADGLHVDEENEREVD 197
E +E G+ K D+DGEE++ ++ + D
Sbjct: 737 ------EKKEKGKEDK--------------------DKDGEEKKSKKDKEKKKDKNEDDD 838
Query: 198 DGNEETELKYDKGY 211
G E KGY
Sbjct: 839 GG*ERRG*SICKGY 880
Score = 29.3 bits (64), Expect = 2.4
Identities = 18/73 (24%), Positives = 36/73 (48%)
Frame = +2
Query: 4 EKRMKNENTRCKMKNEKPFSSDLESGLRDRRDGDGEEDREKVKVNEEKEVDHGKKLEMWN 63
+K +E K K +K E+ + DGD ++D+EK K ++++ K +
Sbjct: 605 DKTDVDEGKDKKDKEKKKKEKKEENVKGEEEDGDEKKDKEKKKKEKKEKGKEDKDKDGEE 784
Query: 64 KMDYKERKKIREK 76
K K+++K ++K
Sbjct: 785 KKSKKDKEKKKDK 823
>TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~unknown
protein {Arabidopsis thaliana}, partial (46%)
Length = 2077
Score = 50.4 bits (119), Expect = 1e-06
Identities = 60/230 (26%), Positives = 96/230 (41%), Gaps = 15/230 (6%)
Frame = +2
Query: 53 VDHGKKLEMWNKMDYKERKKIR------------EKMVRKCRRAGLDGDGEKEREAGEDL 100
+D GKKLE +++ +E + E + +K + G +G+ + E E ED
Sbjct: 329 LDVGKKLEKKKRVELEEEESDDFDEELDDDDDDFEGVEKKKAKGGSEGEDDFEEEDDED- 505
Query: 101 ESGRRDGLDGYGEEEREIGEVDEEKEVKLGKKFGMAWIQVNHYELREHGERRKCGLLEFN 160
E G D D E+E+ G E+K +K+ + + ++YE E
Sbjct: 506 EEGSEDEDDEEDEKEKVKGGGIEDKFLKIDELTEYLEKEEDNYEKGEE------------ 649
Query: 161 FSIDLDSGIRDEIDEDGEEEREADGLHVDEENEREVDDGNEETELKYDKGYEMVRILVNH 220
RDE DED EE+ E + E ++ + DD +EE E D G
Sbjct: 650 ---------RDEADEDSEEDDELEKAGEFEMDDEDDDDDDEEDEEAEDMG---------- 772
Query: 221 NVLKEQYMDKYKQEKRYNSKNLIVEYDG-LDRVE--EEEEREVGEVEKDR 267
NV E + K EK K+ ++E G D +E ++++R EK R
Sbjct: 773 NVRYEDFFGG-KNEKGSKRKDQLLEVSGDEDDMESTKQKKRTASTYEKQR 919
Score = 30.8 bits (68), Expect = 0.82
Identities = 33/110 (30%), Positives = 54/110 (49%), Gaps = 9/110 (8%)
Frame = +2
Query: 251 RVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGLD 310
++E+++ E+ E E D E +++ + E+K+ K G E E + E D
Sbjct: 344 KLEKKKRVELEEEESDDFDEELDDDDDDFEGVEKKKAKGGSEGEDDFEEED--------D 499
Query: 311 GDEEEER-EGDEEKEVKHVK----EKK--KIEKLEK--ENEEKKVKRGKD 351
DEE E DEE E + VK E K KI++L + E EE ++G++
Sbjct: 500 EDEEGSEDEDDEEDEKEKVKGGGIEDKFLKIDELTEYLEKEEDNYEKGEE 649
>AJ499220 similar to SP|Q09863|YAF Hypothetical protein C29E6.10c in
chromosome I. [Fission yeast] {Schizosaccharomyces
pombe}, partial (12%)
Length = 518
Score = 47.4 bits (111), Expect = 8e-06
Identities = 44/178 (24%), Positives = 86/178 (47%)
Frame = -3
Query: 171 DEIDEDGEEEREADGLHVDEENEREVDDGNEETELKYDKGYEMVRILVNHNVLKEQYMDK 230
DE +EDGEE+ + + E+ +++G ++ + +E R+L Y +K
Sbjct: 420 DEYEEDGEEDNQTE--------EQRMEEGRRMFQIFAARMFEQ-RVL-------NAYREK 289
Query: 231 YKQEKRYNSKNLIVEYDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHG 290
QE++ L+ +EEE +RL +EERE+ + +EKE K
Sbjct: 288 VAQERQQKL---------LEELEEE----------NRL----KEERELKKL-KEKERKKA 181
Query: 291 KELEMNISRELESGIRDGLDGDEEEEREGDEEKEVKHVKEKKKIEKLEKENEEKKVKR 348
K + +E E R+ EE+ + + EK+++ +++++ E+L+KE E ++ K+
Sbjct: 180 KNRALKQQKEEERARREAERLAEEKAQRAEREKKLEEERKRREEERLKKEAERRQRKK 7
Score = 41.2 bits (95), Expect = 6e-04
Identities = 48/178 (26%), Positives = 83/178 (45%)
Frame = -3
Query: 171 DEIDEDGEEEREADGLHVDEENEREVDDGNEETELKYDKGYEMVRILVNHNVLKEQYMDK 230
DE +++ E+E E DG ++ E+ +++G ++ + +E R+L Y +K
Sbjct: 444 DEDEDEDEDEYEEDGEEDNQTEEQRMEEGRRMFQIFAARMFEQ-RVL-------NAYREK 289
Query: 231 YKQEKRYNSKNLIVEYDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHG 290
QE++ L+ +EEE +RL +EERE+ + +EKE K
Sbjct: 288 VAQERQQKL---------LEELEEE----------NRL----KEERELKKL-KEKERKKA 181
Query: 291 KELEMNISRELESGIRDGLDGDEEEEREGDEEKEVKHVKEKKKIEKLEKENEEKKVKR 348
K + +E E R E ER EEK + +E KK+E+ K EE+++K+
Sbjct: 180 KNRALKQQKEEERARR-------EAERLA-EEKAQRAERE-KKLEEERKRREEERLKK 34
Score = 36.2 bits (82), Expect = 0.019
Identities = 33/120 (27%), Positives = 57/120 (47%), Gaps = 20/120 (16%)
Frame = -3
Query: 249 LDRVEEEEEREVGEV---EKDRLHGEEEEEREVSVVNEEKELKHGKEL------------ 293
+ R E+E + GE ++D E EE+ E EE+ ++ G+ +
Sbjct: 492 MQREEDESAMDRGEYYDEDEDEDEDEYEEDGEEDNQTEEQRMEEGRRMFQIFAARMFEQR 313
Query: 294 EMNISRELESGIRDGLDGDE-EEEREGDEEKEVKHVKEKK----KIEKLEKENEEKKVKR 348
+N RE + R +E EEE EE+E+K +KEK+ K L+++ EE++ +R
Sbjct: 312 VLNAYREKVAQERQQKLLEELEEENRLKEERELKKLKEKERKKAKNRALKQQKEEERARR 133
>TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein T28I19.100
- Arabidopsis thaliana, partial (14%)
Length = 1460
Score = 46.2 bits (108), Expect = 2e-05
Identities = 78/329 (23%), Positives = 127/329 (37%), Gaps = 6/329 (1%)
Frame = +3
Query: 4 EKRMKNENTRCKMKNEKPFSSDLESGLRDRRDGDGEEDREKVKVNEEKEVDHGKKLEMWN 63
+K +T+ ++ E L+ G +D G E D+ + +EE+E D M N
Sbjct: 279 KKEFDKNDTKLPIRTET--DQILKLGRKDLHPGKVEADKNEG--HEEEEEDEHIVYNMQN 446
Query: 64 KMDYKERKKIREKMVRKCRRAGLDGDGEKEREAGEDLESGRRDGLDGYGEEEREIGEVDE 123
K ++ E+++ G +G+ + E ED RR+ D EE + EV E
Sbjct: 447 KREHDEQQQ-----------EGEEGNKHETEEESEDNVHERREEQDE--EENKHGAEVQE 587
Query: 124 EKEVKLGKKFGMAWIQVNHYELREHGERRKCGLLEFNFSIDLDSGIRDEIDEDGEEEREA 183
E E K E E D G EIDE+ E+ EA
Sbjct: 588 ENESK-----------------SEEVE---------------DEGGDVEIDENDHEKSEA 671
Query: 184 DGLH----VDEENEREVDDGNEETELKYDKGYEMVRILVNHNVLKEQYMDKYKQEKRYNS 239
D VDEE ++E ++G++ETE DK E LV ++ E + YK + ++
Sbjct: 672 DNDREDEVVDEEKDKE-EEGDDETE-NEDKEDEEKGGLVENHENHEAREEHYKADDASSA 845
Query: 240 KNLIVEYDGLDRVEEEEEREVGEVEKD--RLHGEEEEEREVSVVNEEKELKHGKELEMNI 297
V +D E E G +E L + E E + +E ++ +L++
Sbjct: 846 ----VAHD-----THETSTETGNLEHSDLSLQNTRKPENETNHSDESYGSQNVSDLKVT- 995
Query: 298 SRELESGIRDGLDGDEEEEREGDEEKEVK 326
EL G+ +E + + K
Sbjct: 996 EGELTDGVSSNATAGKETGNDSFSNETAK 1082
>TC88509 similar to PIR|T01826|T01826 microfibril-associated protein homolog
T15F16.8 - Arabidopsis thaliana, partial (83%)
Length = 1306
Score = 45.4 bits (106), Expect = 3e-05
Identities = 43/162 (26%), Positives = 77/162 (46%), Gaps = 7/162 (4%)
Frame = +1
Query: 189 DEENEREVDDGNEETELKYDKGYEMVRILVNHNVLKEQYMDKYKQEKRYNSKNLIVEYDG 248
+EE E E ++ EE++ + D E + + +K ++ K + ++ I E +
Sbjct: 505 EEEEEEEEEEEEEESDYETDSDEEYTGVAM----VKPVFVPK-------SERDTIAERER 651
Query: 249 LDRVEEEEEREVGEVEKDRLHGEEEEEREVSV--VNEEKELKHGKELEMNISRELESGIR 306
L E EE + E K R+ E R++ V + +++E++ ELE NI
Sbjct: 652 L----EAEEEALEEARKRRMEERRNETRQIVVEEIRKDQEIQKNIELEANI--------- 792
Query: 307 DGLDGDEE----EEREGDEEKEVKHVK-EKKKIEKLEKENEE 343
D +D D+E EE E + +E+ +K +++ E L KE EE
Sbjct: 793 DDVDTDDELNEAEEYEAWKVREIGRIKRDREDREALLKEKEE 918
>AJ499968 weakly similar to GP|23495961|gb| hypothetical protein {Plasmodium
falciparum 3D7}, partial (19%)
Length = 342
Score = 45.1 bits (105), Expect = 4e-05
Identities = 31/88 (35%), Positives = 49/88 (55%)
Frame = -3
Query: 264 EKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGLDGDEEEEREGDEEK 323
E++R EE EE+E + +EKE + KE E ELE R + +E E +E +E +
Sbjct: 331 ERERKEREEREEQER--IAKEKEEQERKEREERERLELE---RIAKEKEERERKEKEERE 167
Query: 324 EVKHVKEKKKIEKLEKENEEKKVKRGKD 351
E + ++ +IEK KE EEK +K ++
Sbjct: 166 ERERLEAVVRIEKERKEAEEKALKEQEE 83
Score = 33.9 bits (76), Expect = 0.097
Identities = 23/63 (36%), Positives = 35/63 (55%), Gaps = 1/63 (1%)
Frame = -3
Query: 284 EKELKHGKELEMNISRELESGIRDGLDGDEEEEREGDEEKEVKHV-KEKKKIEKLEKENE 342
EKE + KE E RE + I + E +ERE E E++ + KEK++ E+ EKE
Sbjct: 340 EKEERERKERE---EREEQERIAKEKEEQERKEREERERLELERIAKEKEERERKEKEER 170
Query: 343 EKK 345
E++
Sbjct: 169 EER 161
>CB892018
Length = 389
Score = 44.7 bits (104), Expect = 5e-05
Identities = 31/112 (27%), Positives = 58/112 (51%)
Frame = +1
Query: 4 EKRMKNENTRCKMKNEKPFSSDLESGLRDRRDGDGEEDREKVKVNEEKEVDHGKKLEMWN 63
+K +++E+ R + ++ K D +G + RD D +R+ ++ +E H ++ + +
Sbjct: 91 KKSLRDEDVRGERRHGK--DEDYPTGRKHLRDEDDHGERKIRRIEDE----HAERKSLRD 252
Query: 64 KMDYKERKKIREKMVRKCRRAGLDGDGEKEREAGEDLESGRRDGLDGYGEEE 115
DY ERK +R+ + + R+ LDGD E+ + RDG D YGE +
Sbjct: 253 GDDYGERKNLRDDVGSRERKNSLDGDVRGEKRSS-------RDG-DDYGERK 384
Score = 32.7 bits (73), Expect = 0.22
Identities = 40/159 (25%), Positives = 67/159 (41%), Gaps = 3/159 (1%)
Frame = +1
Query: 40 EDREKVKVNEEKEVDHGKKLEMWNKMDYKERKKIREKMVRKCRRAGLDGDGEKEREAGED 99
+D + K + E D ++ D +E+K +R++ VR RR G D ED
Sbjct: 1 QDNHEEKKHGRNEDDSRDGKHKRDEEDRREKKSLRDEDVRGERRHGKD----------ED 150
Query: 100 LESGRRDGLDGYGEEEREIGEVDEEKEVKLGKKFGMAWIQVNHYELREHGERRKCGLLEF 159
+GR+ D ER+I +++E + + G ++GER+
Sbjct: 151 YPTGRKHLRDEDDHGERKIRRIEDEHAERKSLRDG-----------DDYGERK------- 276
Query: 160 NFSIDLDSGIR-DEIDED--GEEEREADGLHVDEENERE 195
N D+ S R + +D D GE+ DG D+ ER+
Sbjct: 277 NLRDDVGSRERKNSLDGDVRGEKRSSRDG---DDYGERK 384
Score = 31.6 bits (70), Expect = 0.48
Identities = 30/125 (24%), Positives = 58/125 (46%), Gaps = 31/125 (24%)
Frame = +1
Query: 234 EKRYNSKNLIVEYDGLDRVEEEEEREVG-----EVEKDRLHGEEEE-------------- 274
E++ + +N DG + +EE+ RE +V +R HG++E+
Sbjct: 13 EEKKHGRNEDDSRDGKHKRDEEDRREKKSLRDEDVRGERRHGKDEDYPTGRKHLRDEDDH 192
Query: 275 -EREVSVVNEE----KELKHG------KELEMNI-SRELESGIRDGLDGDEEEEREGDEE 322
ER++ + +E K L+ G K L ++ SRE ++ + + G++ R+GD+
Sbjct: 193 GERKIRRIEDEHAERKSLRDGDDYGERKNLRDDVGSRERKNSLDGDVRGEKRSSRDGDDY 372
Query: 323 KEVKH 327
E K+
Sbjct: 373 GERKN 387
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 43.1 bits (100), Expect = 2e-04
Identities = 29/96 (30%), Positives = 46/96 (47%)
Frame = +3
Query: 253 EEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGLDGD 312
E EE + GEVE++ +EEEE E EE+E +
Sbjct: 48 EHNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEE-------------------------E 152
Query: 313 EEEEREGDEEKEVKHVKEKKKIEKLEKENEEKKVKR 348
EEEE E D+E+E + +E+++ E ++E EE ++ R
Sbjct: 153 EEEEDEHDDEEEEEEEEEEEEEEDDDEEGEEDEIDR 260
Score = 37.4 bits (85), Expect = 0.009
Identities = 28/64 (43%), Positives = 35/64 (53%)
Frame = +3
Query: 252 VEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGLDG 311
VEEEE+ E E E+D H EEEEE E EE+E +H E E E E D +G
Sbjct: 78 VEEEED-EHDEEEEDE-HDEEEEEEE----EEEEEDEHDDEEEEEEEEEEEEEEDDDEEG 239
Query: 312 DEEE 315
+E+E
Sbjct: 240 EEDE 251
Score = 36.6 bits (83), Expect = 0.015
Identities = 20/47 (42%), Positives = 27/47 (56%)
Frame = +3
Query: 250 DRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELEMN 296
D +EEEE E E E+D EEEEE E EE + + G+E E++
Sbjct: 117 DEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEEDDDEEGEEDEID 257
Score = 36.2 bits (82), Expect = 0.019
Identities = 31/112 (27%), Positives = 50/112 (43%)
Frame = +3
Query: 170 RDEIDEDGEEEREADGLHVDEENEREVDDGNEETELKYDKGYEMVRILVNHNVLKEQYMD 229
R +++E EEE E E E E D+ +EE E ++D+ +E+ +
Sbjct: 33 RRKVEEHNEEEDEG-------EVEEEEDEHDEEEEDEHDE--------------EEEEEE 149
Query: 230 KYKQEKRYNSKNLIVEYDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVV 281
+ ++E ++ D EEEEE E E E D GEE+E +S +
Sbjct: 150 EEEEEDEHD-----------DEEEEEEEEEEEEEEDDDEEGEEDEIDRISTL 272
Score = 32.7 bits (73), Expect = 0.22
Identities = 25/75 (33%), Positives = 34/75 (45%), Gaps = 2/75 (2%)
Frame = +3
Query: 132 KFGMAWIQVNHYELREHGERRKCGLLEFNFSIDLDSGIRDEIDEDGEEEREAD--GLHVD 189
K MA ++ EH E G +E + D DE DE+ EEE E + H D
Sbjct: 3 KMTMAESSWKRRKVEEHNEEEDEGEVEEEED-EHDEEEEDEHDEEEEEEEEEEEEDEHDD 179
Query: 190 EENEREVDDGNEETE 204
EE E E ++ EE +
Sbjct: 180 EEEEEEEEEEEEEED 224
>TC89483 similar to GP|18377662|gb|AAL66981.1 unknown protein {Arabidopsis
thaliana}, partial (35%)
Length = 1711
Score = 42.7 bits (99), Expect = 2e-04
Identities = 44/170 (25%), Positives = 80/170 (46%), Gaps = 8/170 (4%)
Frame = +1
Query: 189 DEENEREVDDGNEETELKYDKGYEMVRILVNHNVLKEQYMDKYKQEKRYNSKNLIVEYDG 248
D ++R+ + + E ELK +K E+ +++Y++E E D
Sbjct: 115 DRRHDRKSRERDRERELKREK---------------ERVLERYERE---------AERDR 222
Query: 249 LDRVEEEEEREVGEVEKD---RLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGI 305
+ R E E++R + EVE+ +L E ERE +EKE ++ KE E + R+ I
Sbjct: 223 I-RKEREQKRRIEEVERQFELQLKEWEYRERE-----KEKERQYEKEKEKDRERKRRKEI 384
Query: 306 RDGLDGDEEEERE-----GDEEKEVKHVKEKKKIEKLEKENEEKKVKRGK 350
+ D+ + R+ EEK K ++EK+ + +K+ EE+++ K
Sbjct: 385 LYDEEDDDGDSRKRWRXNAIEEKRNKRLREKED-DLADKQKEEEEIAEAK 531
>AJ500818 weakly similar to GP|10435304|dbj unnamed protein product {Homo
sapiens}, partial (12%)
Length = 457
Score = 41.6 bits (96), Expect = 5e-04
Identities = 25/91 (27%), Positives = 50/91 (54%)
Frame = -3
Query: 250 DRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGL 309
D VEE+ + E + EK + +EE+R+ EEK + ++ E + + R
Sbjct: 275 DPVEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQEEKRQE- 99
Query: 310 DGDEEEEREGDEEKEVKHVKEKKKIEKLEKE 340
+ +EE+R+ ++ +E K ++EK++ EK ++E
Sbjct: 98 EKRQEEKRQEEKRQEEKRLEEKRQEEKRQEE 6
>TC85290 similar to GP|7767653|gb|AAF69150.1| F27F5.2 {Arabidopsis
thaliana}, partial (32%)
Length = 1558
Score = 41.6 bits (96), Expect = 5e-04
Identities = 30/114 (26%), Positives = 55/114 (47%)
Frame = +3
Query: 42 REKVKVNEEKEVDHGKKLEMWNKMDYKERKKIREKMVRKCRRAGLDGDGEKEREAGEDLE 101
+EK K E K + K E + + KE++K +EK ++ R + + KER ++ +
Sbjct: 720 KEKAKEKERKREEEKAKKE--KEREEKEKRKEKEKKEKERER---EKEKSKERHKKDESD 884
Query: 102 SGRRDGLDGYGEEEREIGEVDEEKEVKLGKKFGMAWIQVNHYELREHGERRKCG 155
S +D DG+G E + E D+E++ + + M + E E + R+ G
Sbjct: 885 SDNQDMTDGHGYREEKKKEKDKERKHRRRHQSSMDDVDSEKDEKEESRKSRRHG 1046
Score = 36.2 bits (82), Expect = 0.019
Identities = 31/117 (26%), Positives = 60/117 (50%), Gaps = 14/117 (11%)
Frame = +3
Query: 249 LDRVEEEEEREVGEVE-------------KDRLHGEEEEEREVSVVNEEKELKHGKELEM 295
L+R +E+EE+E + + KD + EE + + ++ + G E
Sbjct: 507 LERAKEKEEKEAKKRQRLADDFTNLLYTLKDIITSSTWEECKALFEDTQEYISIGNE--- 677
Query: 296 NISREL-ESGIRDGLDGDEEEEREGDEEKEVKHVKEKKKIEKLEKENEEKKVKRGKD 351
+ S+E+ E I + +E+ER+ +EEK K + ++K ++ EKE +EK+ +R K+
Sbjct: 678 SYSKEIFEEYITYLKEKAKEKERKREEEKAKKEKEREEKEKRKEKEKKEKEREREKE 848
Score = 30.4 bits (67), Expect = 1.1
Identities = 25/82 (30%), Positives = 42/82 (50%), Gaps = 2/82 (2%)
Frame = +3
Query: 272 EEEEREVSVVNEE--KELKHGKELEMNISRELESGIRDGLDGDEEEEREGDEEKEVKHVK 329
E+ + +S+ NE KE+ + + + R+ +E+ERE EKE + K
Sbjct: 642 EDTQEYISIGNESYSKEIFEEYITYLKEKAKEKERKREEEKAKKEKERE---EKEKRKEK 812
Query: 330 EKKKIEKLEKENEEKKVKRGKD 351
EKK+ E+ E+E E+ K + KD
Sbjct: 813 EKKEKER-EREKEKSKERHKKD 875
>TC87729 similar to GP|9759461|dbj|BAB10377.1 gene_id:MAF19.15~unknown
protein {Arabidopsis thaliana}, partial (37%)
Length = 2210
Score = 41.2 bits (95), Expect = 6e-04
Identities = 69/304 (22%), Positives = 114/304 (36%), Gaps = 19/304 (6%)
Frame = +1
Query: 11 NTRCKMKNEKPFSSDLESGLRDRRDGDGEEDREKVKVNEEKEVDHGKKLEMWNKMDYKER 70
N R K EKPFSS R RR GEE R ++ N ++ +E
Sbjct: 1 NKRSKRCYEKPFSSLFPPKQRQRRKMGGEEKRHQMMQN------------LFGDQSEEEE 144
Query: 71 KKIREKMVRKCRRAGLDGDGEKEREAGEDLESGR------RDGLDGYGEEEREIGEVDEE 124
++ E E E+ L S +DG +G GE E + GEV+EE
Sbjct: 145 DEL-----------------ESEHESNPQLNSDEGEGGVGQDGEEGEGEVEGQ-GEVEEE 270
Query: 125 KEVKLGKKFGMAWIQVNHYELREHGERRKCGLLEFNFSIDLDSGIRDEIDEDGEEEREAD 184
E + G++ +V + E E R D DS +D+ ++ + D
Sbjct: 271 AESE-GER------EVGDQRVEEESEGR---------GEDTDSDAKDDYNQRVVTSKRRD 402
Query: 185 GLHV-DEENEREVD------------DGNEETELKYDKGYEMVRILVNHNVLKEQYMDKY 231
+ +++E+EVD D + T D E+ + + + +E
Sbjct: 403 VVESGSDDDEQEVDAAARRSISGSPRDDKDHTH-DLDSAPEIRDVFGDFDDEEEDMGYAA 579
Query: 232 KQEKRYNSKNLIVEYDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGK 291
+Q+ +S E +G E+ ++ D H E EE + + ++K L
Sbjct: 580 QQDIEQDSNRYPAEEEG----SYEKSLRPEDILADEEHQYESEEENIEMKTKDKPLGPPL 747
Query: 292 ELEM 295
ELE+
Sbjct: 748 ELEI 759
Score = 32.0 bits (71), Expect = 0.37
Identities = 26/97 (26%), Positives = 51/97 (51%), Gaps = 9/97 (9%)
Frame = +1
Query: 264 EKDRLHGEEEEEREVSVV--------NEEKELKHGKELEMNISRELESGI-RDGLDGDEE 314
++ ++ GEE+ + + + +E E +H ++N S E E G+ +DG +G+ E
Sbjct: 64 QRRKMGGEEKRHQMMQNLFGDQSEEEEDELESEHESNPQLN-SDEGEGGVGQDGEEGEGE 240
Query: 315 EEREGDEEKEVKHVKEKKKIEKLEKENEEKKVKRGKD 351
E +G+ E+E + E+ E ++ EE+ RG+D
Sbjct: 241 VEGQGEVEEEAESEGER---EVGDQRVEEESEGRGED 342
Score = 28.1 bits (61), Expect = 5.3
Identities = 21/75 (28%), Positives = 33/75 (44%), Gaps = 6/75 (8%)
Frame = +3
Query: 245 EYDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHG------KELEMNIS 298
EY+ D +EEEER + D E E+E E + +E + K+ N+
Sbjct: 1620 EYETED--DEEEERPRSRMRDDDSEAEYEDEEEDEQIEQENDASDEDEDEGLKQKSKNLK 1793
Query: 299 RELESGIRDGLDGDE 313
R+ + + G D DE
Sbjct: 1794 RKAK---KKGFDSDE 1829
Score = 27.7 bits (60), Expect = 6.9
Identities = 22/98 (22%), Positives = 42/98 (42%)
Frame = +3
Query: 254 EEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGLDGDE 313
+ +VG + +R E E E + EE+E + + + E E DE
Sbjct: 1569 KSSRHQVGYPDDERDESEYETEDD-----EEEERPRSRMRDDDSEAEYE---------DE 1706
Query: 314 EEEREGDEEKEVKHVKEKKKIEKLEKENEEKKVKRGKD 351
EE+ + ++E + E + +++ K + K K+G D
Sbjct: 1707 EEDEQIEQENDASDEDEDEGLKQKSKNLKRKAKKKGFD 1820
>TC86552 similar to PIR|G85436|G85436 hypothetical protein AT4g36980
[imported] - Arabidopsis thaliana, partial (59%)
Length = 1785
Score = 40.4 bits (93), Expect = 0.001
Identities = 35/133 (26%), Positives = 63/133 (47%), Gaps = 3/133 (2%)
Frame = +2
Query: 174 DEDGEEEREADGLHVDEENEREVDDGNEETELKYDKGYEMVRILVNHNVLKE---QYMDK 230
D +G+EE + DEE+E + DD + ++ D+G E+ I + V + YMDK
Sbjct: 674 DGNGKEESQISD-DDDEEDEDDEDDEDFNSDDSNDEGMEI--IAKEYGVKRYGWLVYMDK 844
Query: 231 YKQEKRYNSKNLIVEYDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHG 290
+E++ K +I + ++ +E R+ +VE++R ERE + + + L H
Sbjct: 845 KAKEEQKRQKEIIKGDPAIRKLSRKERRKASQVERER-------EREATRIPGTRVLHHD 1003
Query: 291 KELEMNISRELES 303
E S E+
Sbjct: 1004PYRESRQSPTYEA 1042
>TC77946 homologue to GP|10334491|emb|CAC10207. putative splicing factor
{Cicer arietinum}, complete
Length = 2061
Score = 40.4 bits (93), Expect = 0.001
Identities = 34/101 (33%), Positives = 51/101 (49%), Gaps = 2/101 (1%)
Frame = +3
Query: 250 DRVEEEEEREVGEV-EKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDG 308
+R E ER G E+D G E+ERE V + G++ E + RE E R+
Sbjct: 330 ERDREHRERSSGRHRERDGERGSREKEREKDVE------RGGRDKERDREREKE---RER 482
Query: 309 LDGDEEEERE-GDEEKEVKHVKEKKKIEKLEKENEEKKVKR 348
D + E+ERE D+E+E + E+K+ E+ KE E +R
Sbjct: 483 RDKEREKERERRDKEREKERA*ERKRGERETKEEYESS*ER 605
Score = 39.3 bits (90), Expect = 0.002
Identities = 31/125 (24%), Positives = 58/125 (45%), Gaps = 5/125 (4%)
Frame = +3
Query: 232 KQEKRYNSKNLIVEYDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGK 291
K E+ Y + + DG + + +R G +++ + + +R+ E +E G+
Sbjct: 195 KSERTYRKHGIDDDLDGGE--DRRSKRSRGGDDENGSKKDRDRDRDRERDREHRERSSGR 368
Query: 292 ELEMNISRELESGIRDGLDGDEEEERE-----GDEEKEVKHVKEKKKIEKLEKENEEKKV 346
E RDG G E+ERE G +KE +EK++ E+ +KE E+++
Sbjct: 369 HRE-----------RDGERGSREKEREKDVERGGRDKERDREREKER-ERRDKEREKERE 512
Query: 347 KRGKD 351
+R K+
Sbjct: 513 RRDKE 527
Score = 36.2 bits (82), Expect = 0.019
Identities = 31/110 (28%), Positives = 54/110 (48%)
Frame = +3
Query: 17 KNEKPFSSDLESGLRDRRDGDGEEDREKVKVNEEKEVDHGKKLEMWNKMDYKERKKIREK 76
++++ D E+G + RD D + +R++ E +E G+ E + +E++ REK
Sbjct: 258 RSKRSRGGDDENGSKKDRDRDRDRERDR----EHRERSSGRHRERDGERGSREKE--REK 419
Query: 77 MVRKCRRAGLDGDGEKEREAGEDLESGRRDGLDGYGEEEREIGEVDEEKE 126
V R G D + ++ERE R+ D E+ERE + + EKE
Sbjct: 420 DVE---RGGRDKERDREREK-------ERERRDKEREKERERRDKEREKE 539
Score = 32.0 bits (71), Expect = 0.37
Identities = 24/78 (30%), Positives = 39/78 (49%)
Frame = +3
Query: 19 EKPFSSDLESGLRDRRDGDGEEDREKVKVNEEKEVDHGKKLEMWNKMDYKERKKIREKMV 78
EK D+E G RD+ E DRE+ K E ++ + K+ E +K KER R++
Sbjct: 402 EKEREKDVERGGRDK-----ERDREREKERERRDKEREKERERRDKEREKERA*ERKRGE 566
Query: 79 RKCRRAGLDGDGEKEREA 96
R+ + + E++R A
Sbjct: 567 RETKEE-YESS*ERQRRA 617
>TC78233 similar to GP|15809838|gb|AAL06847.1 AT5g51120/MWD22_6 {Arabidopsis
thaliana}, partial (66%)
Length = 1183
Score = 40.0 bits (92), Expect = 0.001
Identities = 32/101 (31%), Positives = 55/101 (53%), Gaps = 2/101 (1%)
Frame = +3
Query: 246 YDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGI 305
Y G+D+ +RE + EKD+ ++EEE +++ EE E+ +G ++ E++
Sbjct: 3 YAGIDKNLSVRKREREK*EKDKEEKKKEEEDRIAMEQEEHEV-YGADIP---DEEVD--- 161
Query: 306 RDGLDGD-EEEEREGDEEKEVKH-VKEKKKIEKLEKENEEK 344
+D D + E +EGDEE H KE + ++K KE EE+
Sbjct: 162 ---MDADIDAEHQEGDEELASNHTTKELEDMKKRLKEIEEE 275
>BQ141544 similar to GP|7022293|dbj| unnamed protein product {Homo sapiens},
partial (5%)
Length = 1284
Score = 40.0 bits (92), Expect = 0.001
Identities = 62/317 (19%), Positives = 120/317 (37%), Gaps = 16/317 (5%)
Frame = +2
Query: 38 GEEDREKVKVNEEKEVDHGKKLEMWNKMDYKERKKIREKMVRKCRRAGLDGDG-----EK 92
G E K + E GK+ E + + R+ I + R G D G E+
Sbjct: 308 GREGARKRIKKYKNE*GKGKREEREFRHEKDPRRAIGRRATTPGPRRGGDARGRGGRTEE 487
Query: 93 EREAGEDLESGRRDGLDGYGEEEREIGEVDEEKEVKLGKKFGMAWIQVNHYELREHGERR 152
G + R DG GE + E + GK E E G R
Sbjct: 488 GATGGRGVGGRRSDGCMEGGEAPGDC*SKGERGGSRYGK*V----------ERSEVGASR 637
Query: 153 KC--GLLEFNFSIDLDSGIRDEID-EDGEEEREADGLHVDEENEREVDDGN--------E 201
C G+ ++ + + +D E+ + +A+ HVDE ER++DD +
Sbjct: 638 GCEGGVARRRLQVNAHTQAQSRVDDEENTRDGDAEDAHVDE*IERQIDDAKIRARRRR*D 817
Query: 202 ETELKYDKGYEMVRILVNHNVLKEQYMDKYKQEKRYNSKNLIVEYDGLDRVEEEEEREVG 261
++ + + VR+ H V + + + R + V + +R +R
Sbjct: 818 NSKTRASARDDAVRVTKIHAVDIDIER*RA*YDSRRYT*ITTVRWHERERTNTPRDRCAK 997
Query: 262 EVEKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGLDGDEEEEREGDE 321
E + + + R++ V E+ + G+ RE I+ ++ E+E RE +
Sbjct: 998 ECVRCGIREHTQHARDMRVAGRER--REGRRRRRRRVRE-RGKIKKEIE-SEKERRERER 1165
Query: 322 EKEVKHVKEKKKIEKLE 338
+++ +H +++ +++E
Sbjct: 1166KRDARHARQRDTRKQIE 1216
>TC85439 homologue to SP|P36181|HS80_LYCES Heat shock cognate protein 80.
[Tomato] {Lycopersicon esculentum}, complete
Length = 2401
Score = 39.7 bits (91), Expect = 0.002
Identities = 28/82 (34%), Positives = 43/82 (52%), Gaps = 2/82 (2%)
Frame = +2
Query: 272 EEEEREVSVVNEEKEL--KHGKELEMNISRELESGIRDGLDGDEEEEREGDEEKEVKHVK 329
+E++ E K+L KH + + IS +E I + DE+EE + +EE +V+ V
Sbjct: 608 KEDQLEYLEERRLKDLVKKHSEFISYPISLWVEKTIEKEISDDEDEEEKKEEEGKVEEVD 787
Query: 330 EKKKIEKLEKENEEKKVKRGKD 351
E EKE EEKK K+ K+
Sbjct: 788 E-------EKEKEEKKKKKIKE 832
Score = 28.9 bits (63), Expect = 3.1
Identities = 31/110 (28%), Positives = 53/110 (48%), Gaps = 6/110 (5%)
Frame = +2
Query: 189 DEENEREVDDGNEETELKYDKGYEMVRILVNHNVLKEQYMDKYKQEKRYNSKNLI----- 243
DE+ E G T + G + R +LKE ++ Y +E+R K+L+
Sbjct: 503 DEQYVWESQAGGSFTVTRDTTGEALGRGTKITLILKEDQLE-YLEERRL--KDLVKKHSE 673
Query: 244 -VEYDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKE 292
+ Y VE+ E+E+ + E + ++EEE +V V+EEKE + K+
Sbjct: 674 FISYPISLWVEKTIEKEISDDEDE--EEKKEEEGKVEEVDEEKEKEEKKK 817
Score = 28.9 bits (63), Expect = 3.1
Identities = 19/55 (34%), Positives = 31/55 (55%), Gaps = 4/55 (7%)
Frame = +2
Query: 145 LREHGERRKCGLLEFNFSIDLDSGIRDEI--DEDGEEEREADGL--HVDEENERE 195
+++H E + + S+ ++ I EI DED EE++E +G VDEE E+E
Sbjct: 656 VKKHSE-----FISYPISLWVEKTIEKEISDDEDEEEKKEEEGKVEEVDEEKEKE 805
>TC79918 similar to PIR|T06671|T06671 hypothetical protein T17F15.10 -
Arabidopsis thaliana, partial (39%)
Length = 1269
Score = 37.4 bits (85), Expect = 0.009
Identities = 20/76 (26%), Positives = 35/76 (45%)
Frame = +1
Query: 34 RDGDGEEDREKVKVNEEKEVDHGKKLEMWNKMDYKERKKIREKMVRKCRRAGLDGDGEKE 93
RD D + D + + + +VD G+ + K R++ RK RR+ + KE
Sbjct: 472 RDRDRDRDSGRSRSRSDSDVDRGRDRRKRERGREKGRRRSXSVSPRKQRRSEVKSSDVKE 651
Query: 94 REAGEDLESGRRDGLD 109
R G ++ G+ G+D
Sbjct: 652 RGKGSEMRKGKNVGVD 699
Score = 28.1 bits (61), Expect = 5.3
Identities = 35/177 (19%), Positives = 70/177 (38%), Gaps = 8/177 (4%)
Frame = +1
Query: 176 DGEEEREADGLHVDEENEREVDDGNEETELKYDKGYEMVRILVNHNVLKEQYMDKYKQEK 235
D + +R+ D ++ +VD G + K ++G E R + ++++
Sbjct: 469 DRDRDRDRDSGRSRSRSDSDVDRGRDRR--KRERGREKGR--------RRSXSVSPRKQR 618
Query: 236 RYNSKNLIVEYDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELEM 295
R K+ V+ G E + + VG +HG + E + K +L
Sbjct: 619 RSEVKSSDVKERGKGS-EMRKGKNVGVDYAKLIHGYDN-----MAPAERVKAKMKLQLSE 780
Query: 296 NISRELESGI--------RDGLDGDEEEEREGDEEKEVKHVKEKKKIEKLEKENEEK 344
+R+ + G+ +D DEE E D+ VKH+ + + +E + E++
Sbjct: 781 TAARDSDRGVGWERFEFNKDAPLDDEEVEGAEDDASLVKHIGQSFRFSAVESKREDQ 951
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.306 0.133 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,129,331
Number of Sequences: 36976
Number of extensions: 99185
Number of successful extensions: 1494
Number of sequences better than 10.0: 206
Number of HSP's better than 10.0 without gapping: 1004
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1327
length of query: 351
length of database: 9,014,727
effective HSP length: 97
effective length of query: 254
effective length of database: 5,428,055
effective search space: 1378725970
effective search space used: 1378725970
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0315.14


