

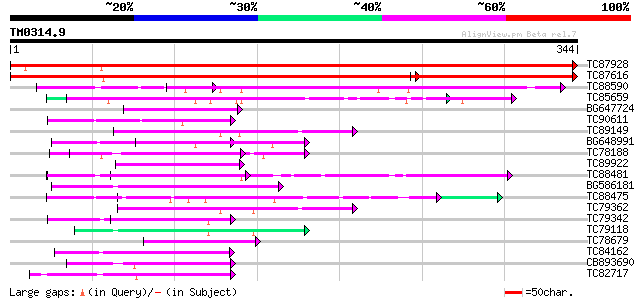
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0314.9
(344 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87928 similar to PIR|A96839|A96839 F23A5.2(form2) [imported] -... 672 0.0
TC87616 similar to PIR|A96839|A96839 F23A5.2(form2) [imported] -... 447 e-180
TC88590 similar to PIR|T52386|T52386 mitotic checkpoint protein ... 160 7e-40
TC85659 SP|O24076|GBLP_MEDSA Guanine nucleotide-binding protein ... 63 2e-10
BG647724 similar to GP|11141605|gb LEUNIG {Arabidopsis thaliana}... 58 6e-09
TC90611 similar to GP|10177223|dbj|BAB10298. contains similarity... 56 2e-08
TC89149 similar to GP|20466231|gb|AAM20433.1 cell cycle switch p... 55 5e-08
BG648991 similar to PIR|C86239|C86 protein T10O24.21 [imported] ... 54 9e-08
TC78188 similar to PIR|C84870|C84870 probable splicing factor [i... 54 1e-07
TC89922 weakly similar to GP|15912305|gb|AAL08286.1 AT4g18900/F1... 52 4e-07
TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclea... 50 1e-06
BG586181 similar to GP|4689316|gb| peroxisomal targeting signal ... 49 3e-06
TC88475 similar to GP|20260442|gb|AAM13119.1 unknown protein {Ar... 48 5e-06
TC79362 WD-repeat cell cycle regulatory protein 48 6e-06
TC79342 similar to GP|10177096|dbj|BAB10430. Notchless protein h... 47 8e-06
TC79118 similar to GP|16323188|gb|AAL15328.1 AT5g67320/K8K14_4 {... 47 1e-05
TC78679 similar to GP|20466538|gb|AAM20586.1 putative WD-repeat ... 46 2e-05
TC84162 similar to GP|15810485|gb|AAL07130.1 putative WD40-repea... 45 4e-05
CB893690 weakly similar to GP|9759025|dbj gene_id:K6A12.9~simila... 44 9e-05
TC82717 weakly similar to GP|21539583|gb|AAM53344.1 putative coa... 43 2e-04
>TC87928 similar to PIR|A96839|A96839 F23A5.2(form2) [imported] -
Arabidopsis thaliana, partial (95%)
Length = 1519
Score = 672 bits (1734), Expect = 0.0
Identities = 318/351 (90%), Positives = 331/351 (93%), Gaps = 7/351 (1%)
Frame = +2
Query: 1 MSTFLSNT---NQNPNKSFEVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVAR--- 54
MS FLSN N NPNKS EVNQPP+DSVSSL FSPK+NLLVATSWDNQVRCWEVAR
Sbjct: 161 MSNFLSNNTNPNPNPNKSIEVNQPPTDSVSSLNFSPKSNLLVATSWDNQVRCWEVARDGA 340
Query: 55 -NVATAPKASISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVK 113
NVAT PKASI+HD PVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQP+TVAMHDAP+K
Sbjct: 341 NNVATMPKASIAHDHPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPITVAMHDAPIK 520
Query: 114 DIAWIPEMSLLVTGSWDRTIKYWDTRQPNPVHTQQLPERCYAMSVKHPLMVVGTADRNIL 173
DIAWIPEM+LL TGSWD+ IKYWDTRQPNPVHTQQLPERCYAM+VKHPLMVVGTADRNI+
Sbjct: 521 DIAWIPEMNLLATGSWDKNIKYWDTRQPNPVHTQQLPERCYAMTVKHPLMVVGTADRNII 700
Query: 174 VYNLQNPQVEFKRIISPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQQGKNFTFK 233
VYNLQNPQVEFKRI+SPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQQ KNFTFK
Sbjct: 701 VYNLQNPQVEFKRIVSPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQQAKNFTFK 880
Query: 234 CHREGNEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNN 293
CHREGNEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQP+PCSAFNN
Sbjct: 881 CHREGNEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPVPCSAFNN 1060
Query: 294 DGSIFAYSVCYDWSKGAENHNPTTAKTNIYLHLPQESEVKGKPRIGATGRK 344
DGSIFAYSVCYDWSKGAENHNP TAK I+LHLPQESEVKGKPRIGATGR+
Sbjct: 1061DGSIFAYSVCYDWSKGAENHNPATAKPYIFLHLPQESEVKGKPRIGATGRR 1213
>TC87616 similar to PIR|A96839|A96839 F23A5.2(form2) [imported] -
Arabidopsis thaliana, partial (96%)
Length = 1471
Score = 447 bits (1151), Expect(2) = e-180
Identities = 208/253 (82%), Positives = 231/253 (91%), Gaps = 4/253 (1%)
Frame = +3
Query: 1 MSTF-LSNTNQNPNKSFEVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARN---V 56
MS+F +NTN NPNKS+EV+QPP+DS+SSL FSPKAN LVATSWDNQVRCWE+A+N V
Sbjct: 129 MSSFGAANTNTNPNKSYEVSQPPTDSISSLSFSPKANFLVATSWDNQVRCWEIAKNGTVV 308
Query: 57 ATAPKASISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIA 116
+ PKASISHDQPVLCSAWKDDGTTVFSGGCDKQ KMWPLLSGGQP+TVAMHDAP+K+IA
Sbjct: 309 TSTPKASISHDQPVLCSAWKDDGTTVFSGGCDKQAKMWPLLSGGQPVTVAMHDAPIKEIA 488
Query: 117 WIPEMSLLVTGSWDRTIKYWDTRQPNPVHTQQLPERCYAMSVKHPLMVVGTADRNILVYN 176
WIPEMSLL TGS D+T+KYWDTRQ NPVHTQQLP+RCY MSV+HPLMVVGTADRN++V+N
Sbjct: 489 WIPEMSLLATGSLDKTVKYWDTRQSNPVHTQQLPDRCYTMSVRHPLMVVGTADRNLIVFN 668
Query: 177 LQNPQVEFKRIISPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHR 236
LQNPQ E+KRI+SPLKYQTRC+AAFPDQQGFLVGSIEGRVGVHHLDD+QQ KNFTFKCHR
Sbjct: 669 LQNPQTEYKRIVSPLKYQTRCVAAFPDQQGFLVGSIEGRVGVHHLDDAQQSKNFTFKCHR 848
Query: 237 EGNEIYSVNSLNF 249
E NEIYS L F
Sbjct: 849 ESNEIYSGQLLKF 887
Score = 201 bits (511), Expect(2) = e-180
Identities = 90/101 (89%), Positives = 95/101 (93%)
Frame = +1
Query: 244 VNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNNDGSIFAYSVC 303
VNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAM RCSQPIPC FNNDGSI+AY+VC
Sbjct: 871 VNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMQRCSQPIPCGTFNNDGSIYAYAVC 1050
Query: 304 YDWSKGAENHNPTTAKTNIYLHLPQESEVKGKPRIGATGRK 344
YDWSKGAENHNPTTAK IY+HLPQESEVKGKPR G+TGR+
Sbjct: 1051YDWSKGAENHNPTTAKNYIYMHLPQESEVKGKPRAGSTGRR 1173
Score = 28.9 bits (63), Expect = 3.0
Identities = 16/59 (27%), Positives = 27/59 (45%), Gaps = 5/59 (8%)
Frame = +3
Query: 243 SVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRL-----KAMLRCSQPIPCSAFNNDGS 296
S++SL+F P + D W+ + KA + QP+ CSA+ +DG+
Sbjct: 204 SISSLSFSPKANFLVATSWDNQVRCWEIAKNGTVVTSTPKASISHDQPVLCSAWKDDGT 380
>TC88590 similar to PIR|T52386|T52386 mitotic checkpoint protein [imported]
- Arabidopsis thaliana, complete
Length = 1395
Score = 160 bits (405), Expect = 7e-40
Identities = 96/267 (35%), Positives = 143/267 (52%), Gaps = 25/267 (9%)
Frame = +2
Query: 96 LLSGGQPMTV--AMHDAPVKDIAWIPEMSLLVT------------GSWDRTIKYWDTR-- 139
L+S P+T+ A PVK I W M L V GSWD+TIK WD R
Sbjct: 302 LVSVPPPITLSDASFSLPVKRIFWESMMLLFVVLSTLTPQGN*SPGSWDKTIKCWDPRGA 481
Query: 140 ---QPNPVHTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQNPQVEFKRIISPLKYQTR 196
+ V T PER Y++S+ +VV TA R++ VY+++N +R S LKYQTR
Sbjct: 482 SGQERTLVGTYAQPERVYSLSLVGHRLVVATAGRHVNVYDMRNMSCPEQRRESSLKYQTR 661
Query: 197 CLAAFPDQQGFLVGSIEGRVGVHHLD--DSQQGKNFTFKCHREGNE----IYSVNSLNFH 250
C+ +P+ G+ + S+EGRV + D ++ Q K + FKCHR+ +Y VN++ FH
Sbjct: 662 CVRCYPNGTGYALSSVEGRVAMEFFDLSEASQAKKYAFKCHRKSEAGRDIVYPVNAMAFH 841
Query: 251 PVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNNDGSIFAYSVCYDWSKGA 310
P++ TFAT G DG N WD ++K+RL + + +F+ DG + A + Y + +G
Sbjct: 842 PIYGTFATGGCDGFVNVWDGNNKKRLYQYSKYPSSVAALSFSRDGRLLAVASSYTFEEGP 1021
Query: 311 ENHNPTTAKTNIYLHLPQESEVKGKPR 337
+ H+ + IY+ E EVK KP+
Sbjct: 1022KPHD----QDAIYVRSVNEIEVKPKPK 1090
Score = 52.4 bits (124), Expect = 3e-07
Identities = 33/110 (30%), Positives = 56/110 (50%)
Frame = +1
Query: 17 EVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWK 76
E+ PPSD +S++ FS ++ L+ +SWD VR ++ A + H PVL +
Sbjct: 121 ELTNPPSDGISNIRFSNHSDHLLVSSWDKTVRLYDA---TADFLRGEFLHGGPVLDCCFH 291
Query: 77 DDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVT 126
DD ++ FS D V+ + + G+ + HDAPV+ + + L+T
Sbjct: 292 DD-SSGFSASADNTVRRL-IFATGKEDILGKHDAPVRCVEYSYAAGQLIT 435
>TC85659 SP|O24076|GBLP_MEDSA Guanine nucleotide-binding protein beta
subunit-like protein. [Alfalfa] {Medicago sativa},
complete
Length = 1261
Score = 62.8 bits (151), Expect = 2e-10
Identities = 67/286 (23%), Positives = 116/286 (40%), Gaps = 13/286 (4%)
Frame = +2
Query: 35 ANLLVATSWDNQVRCWEVARNVAT--APKASIS-HDQPVLCSAWKDDGTTVFSGGCDKQV 91
++++V S D + W + + T P+ ++ H V DG SG D ++
Sbjct: 143 SDMIVTASRDKSIILWHLTKEDKTYGVPRRRLTGHSHFVQDVVLSSDGQFALSGSWDGEL 322
Query: 92 KMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTR-------QPNPV 144
++W L +G H V +A+ + +V+ S DRTIK W+T Q
Sbjct: 323 RLWDLNAGTSARRFVGHTKDVLSVAFSIDNRQIVSASRDRTIKLWNTLGECKYTIQDGDA 502
Query: 145 HTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQNPQVEFKRIISPLKYQTRCLAAFPDQ 204
H+ + ++ S P +V + DR + V+NL N + + ++ +A PD
Sbjct: 503 HSDWVSCVRFSPSTLQPTIVSASWDRTVKVWNLTN--CKLRNTLAGHSGYVNTVAVSPD- 673
Query: 205 QGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHREGNEIYSVNSLNFHPVHHTFATAGSDGA 264
G L S G+ GV L D +GK + I +++L F P + + A ++ +
Sbjct: 674 -GSLCAS-GGKDGVILLWDLAEGKRL---YSLDAGSI--IHALCFSP-NRYWLCAATESS 829
Query: 265 FNFWDKDSK---QRLKAMLRCSQPIPCSAFNNDGSIFAYSVCYDWS 307
WD +SK + LK L+ Y +WS
Sbjct: 830 IKIWDLESKSIVEDLKVDLKTEADAAIGGDTTTKKKVIYCTSLNWS 967
Score = 55.8 bits (133), Expect = 2e-08
Identities = 59/265 (22%), Positives = 103/265 (38%), Gaps = 19/265 (7%)
Frame = +2
Query: 23 SDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTV 82
S V + S ++ SWD ++R W++ N T+ + + H + VL A+ D +
Sbjct: 248 SHFVQDVVLSSDGQFALSGSWDGELRLWDL--NAGTSARRFVGHTKDVLSVAFSIDNRQI 421
Query: 83 FSGGCDKQVKMWPLLSGGQPMTVAMHDAP---VKDIAWIPE--MSLLVTGSWDRTIKYW- 136
S D+ +K+W L G T+ DA V + + P +V+ SWDRT+K W
Sbjct: 422 VSASRDRTIKLWNTL-GECKYTIQDGDAHSDWVSCVRFSPSTLQPTIVSASWDRTVKVWN 598
Query: 137 --DTRQPNPVHTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQNPQVEFKRIISPLKYQ 194
+ + N + A+S L G D IL+++L E KR+ S
Sbjct: 599 LTNCKLRNTLAGHSGYVNTVAVSPDGSLCASGGKDGVILLWDL----AEGKRLYSLDAGS 766
Query: 195 TRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHREGN-----------EIYS 243
F + +L + E + + L+ ++ E + ++
Sbjct: 767 IIHALCFSPNRYWLCAATESSIKIWDLESKSIVEDLKVDLKTEADAAIGGDTTTKKKVIY 946
Query: 244 VNSLNFHPVHHTFATAGSDGAFNFW 268
SLN+ T + +DG W
Sbjct: 947 CTSLNWSADGSTLFSGYTDGVVRVW 1021
>BG647724 similar to GP|11141605|gb LEUNIG {Arabidopsis thaliana}, partial
(25%)
Length = 757
Score = 57.8 bits (138), Expect = 6e-09
Identities = 27/72 (37%), Positives = 40/72 (55%)
Frame = +2
Query: 70 VLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSW 129
V+CS + DG + SGG DK+V +W S Q T+ H + D+ + P M L T S+
Sbjct: 440 VVCSHFSSDGKLLASGGHDKKVVLWYTDSLKQKATLEEHSLLITDVRFSPSMPRLATSSY 619
Query: 130 DRTIKYWDTRQP 141
D+T++ WD P
Sbjct: 620 DKTVRVWDVDNP 655
>TC90611 similar to GP|10177223|dbj|BAB10298. contains similarity to
GTP-binding regulatory protein and WD-repeat
protein~gene_id:MPF21.14, partial (40%)
Length = 855
Score = 56.2 bits (134), Expect = 2e-08
Identities = 33/124 (26%), Positives = 57/124 (45%), Gaps = 10/124 (8%)
Frame = +1
Query: 24 DSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVF 83
D+VSSL S L + SWD ++ W ++ + +HD + DG T +
Sbjct: 484 DTVSSLALSKDGKFLYSVSWDRTIKIWR-TKDFTCLESITNAHDDAINAVVVSYDG-TFY 657
Query: 84 SGGCDKQVKMWPLLSGGQPM----------TVAMHDAPVKDIAWIPEMSLLVTGSWDRTI 133
SG DK++K+W L G T+ H++ + + + S+L +G+ DR+I
Sbjct: 658 SGSADKRIKVWKNLQGDDHNKKKHKHSLVDTLEKHNSGINALVLSSDESILYSGACDRSI 837
Query: 134 KYWD 137
W+
Sbjct: 838 LVWE 849
Score = 30.4 bits (67), Expect = 1.0
Identities = 39/164 (23%), Positives = 62/164 (37%), Gaps = 10/164 (6%)
Frame = +1
Query: 25 SVSSLCFSPKANLLVATSWDNQVRCWEVARN------VATAPKASISHDQPVLCSAWKDD 78
+V SL N L + D ++R W++ + VAT P D
Sbjct: 268 AVKSLVIQSNKNTLFSAHQDRKIRVWKLTNDQTKYTHVATLPTLG-------------DR 408
Query: 79 GTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDT 138
+ + Q++ + T H V +A + L + SWDRTIK W T
Sbjct: 409 ASKILIPKNHVQIRRH------KKCTWVHHVDTVSSLALSKDGKFLYSVSWDRTIKIWRT 570
Query: 139 RQPNPVH--TQQLPERCYAMSVKHP-LMVVGTADRNILVY-NLQ 178
+ + T + A+ V + G+AD+ I V+ NLQ
Sbjct: 571 KDFTCLESITNAHDDAINAVVVSYDGTFYSGSADKRIKVWKNLQ 702
>TC89149 similar to GP|20466231|gb|AAM20433.1 cell cycle switch protein
{Arabidopsis thaliana}, partial (71%)
Length = 1283
Score = 54.7 bits (130), Expect = 5e-08
Identities = 43/155 (27%), Positives = 69/155 (43%), Gaps = 7/155 (4%)
Frame = +2
Query: 64 ISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSL 123
+ H V W D + SGG D Q+ +W S + + H A VK IAW P S
Sbjct: 536 VGHKSEVCGLKWSCDDRELASGGNDNQLLVWNQHSQQPTLRLTEHTAAVKAIAWSPHQSN 715
Query: 124 LVT---GSWDRTIKYWDT---RQPNPVHT-QQLPERCYAMSVKHPLMVVGTADRNILVYN 176
L+ G+ DR I++W+T Q N V T Q+ ++ +V + G + I+V+
Sbjct: 716 LLVSGGGTADRCIRFWNTTNGHQLNSVDTGSQVCNLAWSKNVNELVSTHGYSQNQIMVW- 892
Query: 177 LQNPQVEFKRIISPLKYQTRCLAAFPDQQGFLVGS 211
+ P + ++ + LA PD Q + G+
Sbjct: 893 -KYPSLAKVATLTGHSMRVLYLAMSPDGQTIVTGA 994
Score = 39.7 bits (91), Expect = 0.002
Identities = 29/126 (23%), Positives = 52/126 (41%), Gaps = 6/126 (4%)
Frame = +2
Query: 25 SVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVFS 84
+V ++ +SP + L+ + RC S+ V AW + + S
Sbjct: 677 AVKAIAWSPHQSNLLVSGGGTADRCIRFWNTTNGHQLNSVDTGSQVCNLAWSKNVNELVS 856
Query: 85 --GGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDT---- 138
G Q+ +W S + T+ H V +A P+ +VTG+ D T+++W+
Sbjct: 857 THGYSQNQIMVWKYPSLAKVATLTGHSMRVLYLAMSPDGQTIVTGAGDETLRFWNVFPSM 1036
Query: 139 RQPNPV 144
+ P PV
Sbjct: 1037KTPAPV 1054
Score = 34.7 bits (78), Expect = 0.055
Identities = 33/144 (22%), Positives = 58/144 (39%), Gaps = 5/144 (3%)
Frame = +2
Query: 71 LCSA-WKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSW 129
+CS W +G+ + G QV++W + T+ H +AW +L +GS
Sbjct: 305 VCSVQWTKEGSFISIGTNGGQVQIWDGTKCKKVRTMGGHQTRTGVLAW--NSRILASGSR 478
Query: 130 DRTIKYWDTRQPNPVHTQQLPERCYAMSVKHPL----MVVGTADRNILVYNLQNPQVEFK 185
DR I D R P+ + + + +K + G D +LV+N + Q +
Sbjct: 479 DRNILQHDMRVPSDFIGKLVGHKSEVCGLKWSCDDRELASGGNDNQLLVWNQHSQQPTLR 658
Query: 186 RIISPLKYQTRCLAAFPDQQGFLV 209
++ + +A P Q LV
Sbjct: 659 --LTEHTAAVKAIAWSPHQSNLLV 724
>BG648991 similar to PIR|C86239|C86 protein T10O24.21 [imported] -
Arabidopsis thaliana, partial (35%)
Length = 702
Score = 53.9 bits (128), Expect = 9e-08
Identities = 28/112 (25%), Positives = 57/112 (50%), Gaps = 6/112 (5%)
Frame = +3
Query: 77 DDGTTVFSGGCDKQVKMWPLLSGGQPMTVAM-HDAPVKDIAWIPEMSLLVTGSWDRTIKY 135
+ G + S G D +VK+W + + G+ M M H V+DI + + + ++ +D+ IKY
Sbjct: 360 NSGHLILSAGMDTKVKIWDVFNTGKCMRTYMGHSKAVRDICFTNDGTKFLSAGYDKNIKY 539
Query: 136 WDTRQPNPVHTQQLPERCYAMSV-----KHPLMVVGTADRNILVYNLQNPQV 182
WDT + T + Y + + K +++ G +D+ I+ +++ Q+
Sbjct: 540 WDTETGQVISTFSTGKIPYVVKLNPDEDKQNVLLAGMSDKKIVQWDMNTGQI 695
Score = 46.6 bits (109), Expect = 1e-05
Identities = 32/115 (27%), Positives = 55/115 (47%), Gaps = 3/115 (2%)
Frame = +3
Query: 26 VSSLCFSPKA-NLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVFS 84
VS++ F P + +L+++ D +V+ W+V N + + H + V + +DGT S
Sbjct: 336 VSAIRFFPNSGHLILSAGMDTKVKIWDVF-NTGKCMRTYMGHSKAVRDICFTNDGTKFLS 512
Query: 85 GGCDKQVKMWPLLSGGQPMTVAMHDAP--VKDIAWIPEMSLLVTGSWDRTIKYWD 137
G DK +K W +G T + P VK + ++L+ G D+ I WD
Sbjct: 513 AGYDKNIKYWDTETGQVISTFSTGKIPYVVKLNPDEDKQNVLLAGMSDKKIVQWD 677
Score = 30.0 bits (66), Expect = 1.4
Identities = 15/89 (16%), Positives = 35/89 (38%)
Frame = +3
Query: 11 NPNKSFEVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPV 70
N K S +V +CF+ ++ +D ++ W+ + ++ V
Sbjct: 423 NTGKCMRTYMGHSKAVRDICFTNDGTKFLSAGYDKNIKYWDTETGQVISTFSTGKIPYVV 602
Query: 71 LCSAWKDDGTTVFSGGCDKQVKMWPLLSG 99
+ +D + +G DK++ W + +G
Sbjct: 603 KLNPDEDKQNVLLAGMSDKKIVQWDMNTG 689
>TC78188 similar to PIR|C84870|C84870 probable splicing factor [imported] -
Arabidopsis thaliana, partial (95%)
Length = 1436
Score = 53.5 bits (127), Expect = 1e-07
Identities = 40/163 (24%), Positives = 72/163 (43%), Gaps = 5/163 (3%)
Frame = +1
Query: 25 SVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVFS 84
+V ++ F+P +++ + S D ++ W V + H VL W DGT + S
Sbjct: 319 AVYTMKFNPTGSVVASGSHDKEIFLWNVHGDCKNFMVLK-GHKNAVLDLHWTSDGTQIIS 495
Query: 85 GGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPE-MSLLVTGSWDRTIKYWDTRQPNP 143
DK +++W +G Q + H + V L+V+GS D T K WD RQ
Sbjct: 496 ASPDKTLRLWDTETGKQIKKMVEHLSYVNSCCPTRRGPPLVVSGSDDGTAKLWDMRQRGS 675
Query: 144 VHTQQLPER----CYAMSVKHPLMVVGTADRNILVYNLQNPQV 182
+ T P++ + S + G D ++ +++L+ +V
Sbjct: 676 IQT--FPDKYQITAVSFSDASDKIYTGGIDNDVKIWDLRKGEV 798
Score = 46.2 bits (108), Expect = 2e-05
Identities = 28/109 (25%), Positives = 49/109 (44%), Gaps = 2/109 (1%)
Frame = +1
Query: 37 LLVATSWDNQVRCWEVAR--NVATAPKASISHDQPVLCSAWKDDGTTVFSGGCDKQVKMW 94
L+V+ S D + W++ + ++ T P + ++ D +++GG D VK+W
Sbjct: 613 LVVSGSDDGTAKLWDMRQRGSIQTFPDK-----YQITAVSFSDASDKIYTGGIDNDVKIW 777
Query: 95 PLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQPNP 143
L G MT+ H + + P+ S L+T D + WD R P
Sbjct: 778 DLRKGEVTMTLQGHQDMITSMQLSPDGSYLLTNGMDCKLCIWDMRPYAP 924
Score = 38.1 bits (87), Expect = 0.005
Identities = 27/130 (20%), Positives = 51/130 (38%), Gaps = 12/130 (9%)
Frame = +1
Query: 21 PPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGT 80
P ++++ FS ++ + DN V+ W++ + T H + DG+
Sbjct: 688 PDKYQITAVSFSDASDKIYTGGIDNDVKIWDLRKGEVTMTLQG--HQDMITSMQLSPDGS 861
Query: 81 TVFSGGCDKQVKMWPL------------LSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGS 128
+ + G D ++ +W + L G Q + + W P+ S + GS
Sbjct: 862 YLLTNGMDCKLCIWDMRPYAPQNRCVKILEGHQHN----FEKNLLKCGWSPDGSKVTAGS 1029
Query: 129 WDRTIKYWDT 138
DR + WDT
Sbjct: 1030SDRMVYIWDT 1059
>TC89922 weakly similar to GP|15912305|gb|AAL08286.1 AT4g18900/F13C5_70
{Arabidopsis thaliana}, partial (48%)
Length = 864
Score = 51.6 bits (122), Expect = 4e-07
Identities = 28/80 (35%), Positives = 45/80 (56%), Gaps = 2/80 (2%)
Frame = +1
Query: 65 SHDQPVLCSAW-KDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPE-MS 122
SH VL AW K+ T+ S DK+VK+W +++G +T+ H V+ +AW
Sbjct: 85 SHTDSVLGLAWNKEYSNTLASASADKRVKIWDIVAGKCTITMDHHSDKVQAVAWNHRAQQ 264
Query: 123 LLVTGSWDRTIKYWDTRQPN 142
+L++GS+D T+ D R P+
Sbjct: 265 ILLSGSFDHTVALKDVRTPS 324
Score = 33.9 bits (76), Expect = 0.094
Identities = 24/98 (24%), Positives = 48/98 (48%), Gaps = 2/98 (2%)
Frame = +1
Query: 3 TFLSNTNQNPNKSFEVNQPPSDSVSSLCFSPKA-NLLVATSWDNQVRCWEVARNVATAPK 61
T +SN N +F ++ SV+S+ ++ A NLL S D V+ W+++ N ++
Sbjct: 445 TAMSNATSVQNATFTLHAHDK-SVTSVSYNTAAPNLLATGSMDKTVKLWDLSNNQPSSVA 621
Query: 62 ASISHDQPVLCSAWKDDGTTVFS-GGCDKQVKMWPLLS 98
+ V ++ +D + + GG ++++W LS
Sbjct: 622 SKEPKAGAVFSISFSEDNPFLLAIGGSKGKLQLWDTLS 735
Score = 32.7 bits (73), Expect = 0.21
Identities = 38/148 (25%), Positives = 63/148 (41%), Gaps = 22/148 (14%)
Frame = +1
Query: 23 SDSVSSLCFSPKAN-LLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWK----- 76
SD V ++ ++ +A +L++ S+D+ V +V + S+S D L AW
Sbjct: 220 SDKVQAVAWNHRAQQILLSGSFDHTVALKDVRTPSHSGYTWSVSADVESL--AWDPHTEH 393
Query: 77 ------DDGT-------TVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAW-IPEMS 122
+DGT T S Q T+ HD V +++ +
Sbjct: 394 SFAVSLEDGTIQCFDVRTAMSNATSVQ---------NATFTLHAHDKSVTSVSYNTAAPN 546
Query: 123 LLVTGSWDRTIKYWD--TRQPNPVHTQQ 148
LL TGS D+T+K WD QP+ V +++
Sbjct: 547 LLATGSMDKTVKLWDLSNNQPSSVASKE 630
>TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclear
ribonucleoprotein [imported] - Arabidopsis thaliana,
partial (60%)
Length = 1836
Score = 50.1 bits (118), Expect = 1e-06
Identities = 30/123 (24%), Positives = 52/123 (41%)
Frame = +1
Query: 24 DSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVF 83
+ ++ + F P L S+D R W+V + H + V + DG+
Sbjct: 1204 ERLARIAFHPSGKYLGTASYDKTWRLWDVETEEELLLQEG--HSRSVYGLDFHHDGSLAA 1377
Query: 84 SGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQPNP 143
S G D ++W L +G + + H + I++ P L TG D T + WD R+
Sbjct: 1378 SCGLDALARVWDLRTGRSVLALEGHVKSILGISFSPNGYHLATGGEDNTCRIWDLRKKKS 1557
Query: 144 VHT 146
++T
Sbjct: 1558 LYT 1566
Score = 47.8 bits (112), Expect = 6e-06
Identities = 33/125 (26%), Positives = 54/125 (42%), Gaps = 1/125 (0%)
Frame = +1
Query: 23 SDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTV 82
S SV L F +L + D R W++ + A H + +L ++ +G +
Sbjct: 1327 SRSVYGLDFHHDGSLAASCGLDALARVWDL--RTGRSVLALEGHVKSILGISFSPNGYHL 1500
Query: 83 FSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMS-LLVTGSWDRTIKYWDTRQP 141
+GG D ++W L T+ H + + + P+ LVT S+D T K W +R
Sbjct: 1501 ATGGEDNTCRIWDLRKKKSLYTIPAHSNLISQVKFEPQEGYFLVTASYDMTAKVWSSRDF 1680
Query: 142 NPVHT 146
PV T
Sbjct: 1681 KPVKT 1695
Score = 43.1 bits (100), Expect = 2e-04
Identities = 53/249 (21%), Positives = 101/249 (40%), Gaps = 5/249 (2%)
Frame = +1
Query: 62 ASISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIP-E 120
+ I D+P+ ++ DG + + K+W + + + T+ H D+A+ P
Sbjct: 934 SEIGDDRPLTGCSFSRDGKGLATCSFTGATKLWSMPNVKKVSTLKGHTQRATDVAYSPVH 1113
Query: 121 MSLLVTGSWDRTIKYWDTR----QPNPVHTQQLPERCYAMSVKHPLMVVGTADRNILVYN 176
+ L T S DRT KYW+ + H ++L + S K+ +GTA + +
Sbjct: 1114 KNHLATASADRTAKYWNDQGALLGTFKGHLERLARIAFHPSGKY----LGTASYD-KTWR 1278
Query: 177 LQNPQVEFKRIISPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHR 236
L + + E + ++ + +R + + + G + + D + G++
Sbjct: 1279 LWDVETEEELLLQ--EGHSRSVYGLDFHHDGSLAASCGLDALARVWDLRTGRSV---LAL 1443
Query: 237 EGNEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNNDGS 296
EG+ + S+ ++F P + AT G D WD K+ L + S I F
Sbjct: 1444 EGH-VKSILGISFSPNGYHLATGGEDNTCRIWDLRKKKSLYTIPAHSNLISQVKFEPQEG 1620
Query: 297 IFAYSVCYD 305
F + YD
Sbjct: 1621 YFLVTASYD 1647
Score = 39.7 bits (91), Expect = 0.002
Identities = 30/114 (26%), Positives = 48/114 (41%), Gaps = 2/114 (1%)
Frame = +1
Query: 25 SVSSLCFSPKANLLVATSWDNQVRCWEVAR--NVATAPKASISHDQPVLCSAWKDDGTTV 82
S+ + FSP L DN R W++ + ++ T P S Q +G +
Sbjct: 1459 SILGISFSPNGYHLATGGEDNTCRIWDLRKKKSLYTIPAHSNLISQ---VKFEPQEGYFL 1629
Query: 83 FSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYW 136
+ D K+W T++ H+A V + + + +VT S DRTIK W
Sbjct: 1630 VTASYDMTAKVWSSRDFKPVKTLSGHEAKVTSLDVLGDGGYIVTVSHDRTIKLW 1791
>BG586181 similar to GP|4689316|gb| peroxisomal targeting signal type 2
receptor {Arabidopsis thaliana}, partial (56%)
Length = 804
Score = 48.9 bits (115), Expect = 3e-06
Identities = 40/145 (27%), Positives = 72/145 (49%), Gaps = 4/145 (2%)
Frame = +1
Query: 26 VSSLCFSPK-ANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAW-KDDGTTVF 83
V S ++P+ A++ + S D VR W+V +T + H+ +L W K D +
Sbjct: 46 VYSSVWNPRHADVFASASGDCTVRIWDVREPGSTMILPA--HEHEILSCDWNKYDECIIA 219
Query: 84 SGGCDKQVKMWPLLSGGQPMTVAM-HDAPVKDIAWIPEM-SLLVTGSWDRTIKYWDTRQP 141
+G DK VK+W + S P+ V H V+ + + P + +LLV+ S+D T+ WD
Sbjct: 220 TGSVDKSVKVWDVRSYRVPVAVLNGHGYAVRKVKFSPHVRNLLVSCSYDMTVCVWDFMVE 399
Query: 142 NPVHTQQLPERCYAMSVKHPLMVVG 166
+ + ++ +A+ V ++V G
Sbjct: 400 DALVSRYDHHTEFAVGVDMSVLVEG 474
Score = 39.3 bits (90), Expect = 0.002
Identities = 23/84 (27%), Positives = 37/84 (43%), Gaps = 2/84 (2%)
Frame = +1
Query: 58 TAPKASISHDQPVLCSAWKDDGTTVF-SGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIA 116
T+ + H V S W VF S D V++W + G M + H+ +
Sbjct: 10 TSVRTFKEHAYCVYSSVWNPRHADVFASASGDCTVRIWDVREPGSTMILPAHEHEILSCD 189
Query: 117 WIP-EMSLLVTGSWDRTIKYWDTR 139
W + ++ TGS D+++K WD R
Sbjct: 190 WNKYDECIIATGSVDKSVKVWDVR 261
Score = 30.4 bits (67), Expect = 1.0
Identities = 24/112 (21%), Positives = 49/112 (43%), Gaps = 6/112 (5%)
Frame = +1
Query: 104 TVAMHDAPVKDIAWIPEMS-LLVTGSWDRTIKYWDTRQPN-----PVHTQQLPERCYAMS 157
T H V W P + + + S D T++ WD R+P P H ++ C
Sbjct: 22 TFKEHAYCVYSSVWNPRHADVFASASGDCTVRIWDVREPGSTMILPAHEHEILS-CDWNK 198
Query: 158 VKHPLMVVGTADRNILVYNLQNPQVEFKRIISPLKYQTRCLAAFPDQQGFLV 209
++ G+ D+++ V+++++ +V +++ Y R + P + LV
Sbjct: 199 YDECIIATGSVDKSVKVWDVRSYRVPV-AVLNGHGYAVRKVKFSPHVRNLLV 351
>TC88475 similar to GP|20260442|gb|AAM13119.1 unknown protein {Arabidopsis
thaliana}, partial (96%)
Length = 1315
Score = 48.1 bits (113), Expect = 5e-06
Identities = 61/245 (24%), Positives = 109/245 (43%), Gaps = 5/245 (2%)
Frame = +3
Query: 23 SDSVSSLCFSPKANLLVAT-SWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTT 81
+DSV LC+ PK L+AT S D VR W+ AR+ + +A +S + + +K DGT
Sbjct: 339 TDSVDQLCWDPKHPDLIATASGDKTVRLWD-ARSGKCSQQAELSGEN--INITYKPDGTH 509
Query: 82 VFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAW--IPEMSLLVTGSWD-RTIKYWDT 138
V G D ++ + + +PM + V +IAW EM L TG+ + Y
Sbjct: 510 VAVGNRDDELTILDVRK-FKPMHRRKFNYEVNEIAWNMTGEMFFLTTGNGTVEVLSYPSL 686
Query: 139 RQPNPVHTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQNPQVEFKRIISPLKYQTRCL 198
R + + C A+ VG+AD + ++ + ++ R + L++ R +
Sbjct: 687 RPLDTLMAHTAGCYCIAIDPTGRHFAVGSADSLVSLWVIS--EMLCVRTFTKLEWPVRTI 860
Query: 199 AAFPDQQGFLVGSIEGRVGVHHLDDSQQGKN-FTFKCHREGNEIYSVNSLNFHPVHHTFA 257
+ + G L+ S + + + + GK C ++NS+ ++P ++ A
Sbjct: 861 SF--NHTGDLIASASEDLFI-DISNVHTGKTVHHIPCRA------AMNSVEWNPKYNVLA 1013
Query: 258 TAGSD 262
AG D
Sbjct: 1014YAGDD 1028
Score = 45.4 bits (106), Expect = 3e-05
Identities = 49/241 (20%), Positives = 93/241 (38%), Gaps = 7/241 (2%)
Frame = +3
Query: 66 HDQPVLCSAWKDDGTTVFSGGCDKQVKMWPL--LSGGQPMTVAM--HDAPVKDIAWIPEM 121
H + V AW GT + SG D+ ++W + + G+ + + H V + W P+
Sbjct: 198 HKKKVHSVAWNCIGTKLASGSVDQTARIWHIDPHAHGKVKDIELKGHTDSVDQLCWDPKH 377
Query: 122 -SLLVTGSWDRTIKYWDTRQPNPVHTQQLPERCYAMSVK--HPLMVVGTADRNILVYNLQ 178
L+ T S D+T++ WD R +L ++ K + VG D + + +++
Sbjct: 378 PDLIATASGDKTVRLWDARSGKCSQQAELSGENINITYKPDGTHVAVGNRDDELTILDVR 557
Query: 179 NPQVEFKRIISPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHREG 238
+ +R Y+ +A + F + + G V V L T H G
Sbjct: 558 KFKPMHRR---KFNYEVNEIAWNMTGEMFFLTTGNGTVEV--LSYPSLRPLDTLMAHTAG 722
Query: 239 NEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNNDGSIF 298
+++ P FA +D + W ++ + P+ +FN+ G +
Sbjct: 723 CYCIAID-----PTGRHFAVGSADSLVSLWVISEMLCVRTFTKLEWPVRTISFNHTGDLI 887
Query: 299 A 299
A
Sbjct: 888 A 890
>TC79362 WD-repeat cell cycle regulatory protein
Length = 1800
Score = 47.8 bits (112), Expect = 6e-06
Identities = 38/153 (24%), Positives = 66/153 (42%), Gaps = 7/153 (4%)
Frame = +3
Query: 66 HDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLV 125
H V W D + SGG D ++ +W S + H A VK IAW P + L+
Sbjct: 1035 HKSEVCGLKWSYDNRELASGGNDNKLFVWNQHSTQPVLKYCEHTAAVKAIAWSPHLHGLL 1214
Query: 126 T---GSWDRTIKYWDTRQPNPVHT----QQLPERCYAMSVKHPLMVVGTADRNILVYNLQ 178
G+ DR I++W+T + + Q+ ++ +V + G + I+V+ +
Sbjct: 1215 ASGGGTADRCIRFWNTTTNSHLSCMDTGSQVCNLVWSKNVNELVSTHGYSQNQIIVW--R 1388
Query: 179 NPQVEFKRIISPLKYQTRCLAAFPDQQGFLVGS 211
P + ++ Y+ LA PD Q + G+
Sbjct: 1389 YPTMSKLATLTGHTYRVLYLAISPDGQTIVTGA 1487
Score = 35.8 bits (81), Expect = 0.025
Identities = 31/139 (22%), Positives = 53/139 (37%), Gaps = 12/139 (8%)
Frame = +3
Query: 11 NPNKSFEVNQPPSDSVSSLC----------FSPKANLLVATSWDNQVRCWEVARNVATAP 60
N NK F NQ + V C +SP + L+A+ RC +
Sbjct: 1098 NDNKLFVWNQHSTQPVLKYCEHTAAVKAIAWSPHLHGLLASGGGTADRCIRFWNTTTNSH 1277
Query: 61 KASISHDQPVLCSAWKDDGTTVFS--GGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWI 118
+ + V W + + S G Q+ +W + + T+ H V +A
Sbjct: 1278 LSCMDTGSQVCNLVWSKNVNELVSTHGYSQNQIIVWRYPTMSKLATLTGHTYRVLYLAIS 1457
Query: 119 PEMSLLVTGSWDRTIKYWD 137
P+ +VTG+ D T+ +W+
Sbjct: 1458 PDGQTIVTGAGDETLTFWN 1514
Score = 28.5 bits (62), Expect = 3.9
Identities = 25/102 (24%), Positives = 40/102 (38%)
Frame = +3
Query: 38 LVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLL 97
++A N V W + T + D V W GT + G + +V++W
Sbjct: 705 VLAVGLGNCVYLWNACSSKVTK-LCDLGVDDCVCSVGWAQRGTHLAVGTNNGKVQIWDAA 881
Query: 98 SGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTR 139
+ ++ H V +AW SLL +G D+ I D R
Sbjct: 882 RCKKIRSMEGHRLRVGALAW--SSSLLSSGGRDKNIYQRDIR 1001
>TC79342 similar to GP|10177096|dbj|BAB10430. Notchless protein homolog
{Arabidopsis thaliana}, partial (47%)
Length = 873
Score = 47.4 bits (111), Expect = 8e-06
Identities = 34/120 (28%), Positives = 49/120 (40%), Gaps = 6/120 (5%)
Frame = +3
Query: 24 DSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVF 83
++V S+ FSP L + S D VR W++ T H VLC AW DG +
Sbjct: 498 EAVLSVAFSPDGRQLASGSGDTTVRFWDLGTQ--TPMYTCTGHKNWVLCIAWSPDGKYLV 671
Query: 84 SGGCDKQVKMWPLLSGGQ-PMTVAMHDAPVKDIAWIP-----EMSLLVTGSWDRTIKYWD 137
SG ++ W +G Q + H + I+W P V+ S D + WD
Sbjct: 672 SGSMSGELICWDPQTGKQLGNALTGHKKWITGISWEPVHLNAPCRRFVSASKDGDARIWD 851
Score = 42.7 bits (99), Expect = 2e-04
Identities = 28/77 (36%), Positives = 37/77 (47%), Gaps = 1/77 (1%)
Frame = +3
Query: 62 ASIS-HDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPE 120
A+IS H + VL A+ DG + SG D V+ W L + T H V IAW P+
Sbjct: 477 ATISGHGEAVLSVAFSPDGRQLASGSGDTTVRFWDLGTQTPMYTCTGHKNWVLCIAWSPD 656
Query: 121 MSLLVTGSWDRTIKYWD 137
LV+GS + WD
Sbjct: 657 GKYLVSGSMSGELICWD 707
>TC79118 similar to GP|16323188|gb|AAL15328.1 AT5g67320/K8K14_4 {Arabidopsis
thaliana}, partial (44%)
Length = 1098
Score = 46.6 bits (109), Expect = 1e-05
Identities = 35/155 (22%), Positives = 62/155 (39%), Gaps = 12/155 (7%)
Frame = +2
Query: 40 ATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSG 99
++S DN + ++ N T A H V C W GT + S D K+W +
Sbjct: 242 SSSTDNMIYVCKIGENHPTQTFAG--HQGEVNCVKWDPTGTLLASCSDDITAKIWSVKQD 415
Query: 100 GQPMTVAMHDAPVKDIAWIP---------EMSLLVTGSWDRTIKYWDTRQPNPVHT---Q 147
H + I W P + LL + S+D T+K WD +H+
Sbjct: 416 KYLHDFREHSKEIYTIRWSPTGPGTNNPNKKLLLASASFDSTVKLWDIELGKLIHSLNGH 595
Query: 148 QLPERCYAMSVKHPLMVVGTADRNILVYNLQNPQV 182
+ P A S + G+ D+++ +++L+ ++
Sbjct: 596 RHPVYSVAFSPNGEYIASGSLDKSLHIWSLKEGKI 700
Score = 36.6 bits (83), Expect = 0.015
Identities = 22/80 (27%), Positives = 33/80 (40%)
Frame = +2
Query: 66 HDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLV 125
H P L W+ + T+ S D + + + T A H V + W P +LL
Sbjct: 191 HSGPTLDVDWRTN-TSFASSSTDNMIYVCKIGENHPTQTFAGHQGEVNCVKWDPTGTLLA 367
Query: 126 TGSWDRTIKYWDTRQPNPVH 145
+ S D T K W +Q +H
Sbjct: 368 SCSDDITAKIWSVKQDKYLH 427
>TC78679 similar to GP|20466538|gb|AAM20586.1 putative WD-repeat protein
{Arabidopsis thaliana}, partial (55%)
Length = 1198
Score = 46.2 bits (108), Expect = 2e-05
Identities = 20/71 (28%), Positives = 39/71 (54%)
Frame = +2
Query: 82 VFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQP 141
+++G + V ++ L+SG Q T+ H +PV+D +W P +LV+ SWD + W +
Sbjct: 752 IYTGSHNACVYVYDLVSGAQVATLKHHKSPVRDCSWHPFHPMLVSSSWDGDVVKWQSAGS 931
Query: 142 NPVHTQQLPER 152
+ + + +R
Sbjct: 932 SDMAASSVKKR 964
Score = 33.1 bits (74), Expect = 0.16
Identities = 31/122 (25%), Positives = 54/122 (43%), Gaps = 4/122 (3%)
Frame = +1
Query: 23 SDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTV 82
S + SL FS LVA + + + +++ N + + + D +C A + G +
Sbjct: 184 SFGIFSLKFSTDGKELVAGTSGDSIYVYDLETNKVSLRILAHTADVNTVCFA-DETGHLI 360
Query: 83 FSGGCDKQVKMWP---LLSGGQPMTVAM-HDAPVKDIAWIPEMSLLVTGSWDRTIKYWDT 138
+SG D K+W L + +P V M H + I + ++ D+TIK WD
Sbjct: 361 YSGSDDSFCKVWDRRCLNAKDKPAGVLMVHLEGITFIDSRGDGRYFISNGKDQTIKLWDI 540
Query: 139 RQ 140
R+
Sbjct: 541 RK 546
>TC84162 similar to GP|15810485|gb|AAL07130.1 putative WD40-repeat protein
{Arabidopsis thaliana}, partial (29%)
Length = 628
Score = 45.1 bits (105), Expect = 4e-05
Identities = 31/109 (28%), Positives = 46/109 (41%)
Frame = +1
Query: 28 SLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVFSGGC 87
S+ FSP +V S D +R W ++ + K H VL + + GT + S G
Sbjct: 127 SVEFSPVDQCVVTASGDKTIRIWAISD--GSCLKTFEGHTSSVLRALFVTRGTQIVSCGA 300
Query: 88 DKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYW 136
D VK+W + S T H+ V +A + L TG D + W
Sbjct: 301 DGLVKLWTVKSNECVATYDNHEDKVWALAVGRKTETLATGGSDAVVNLW 447
>CB893690 weakly similar to GP|9759025|dbj gene_id:K6A12.9~similar to unknown
protein~sp|O15736 {Arabidopsis thaliana}, partial (59%)
Length = 916
Score = 43.9 bits (102), Expect = 9e-05
Identities = 26/105 (24%), Positives = 49/105 (45%), Gaps = 2/105 (1%)
Frame = +3
Query: 35 ANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSA--WKDDGTTVFSGGCDKQVK 92
+ L+V+ ++D ++ W++ + T P +C+A + DG T+FSG D ++
Sbjct: 531 SRLVVSAAYDGTIKVWDLMKGYCTNTIMF-----PSICNALCFSTDGQTIFSGHVDGNLR 695
Query: 93 MWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWD 137
+W + SG VA H V I+ ++ +T D +D
Sbjct: 696 LWDIQSGRLLSEVATHSHAVTSISLSRNGNIALTSGGDNLHNLFD 830
Score = 32.3 bits (72), Expect = 0.27
Identities = 23/103 (22%), Positives = 47/103 (45%), Gaps = 2/103 (1%)
Frame = +3
Query: 123 LLVTGSWDRTIKYWDTRQPNPVHTQQLPERCYAM--SVKHPLMVVGTADRNILVYNLQNP 180
L+V+ ++D TIK WD + +T P C A+ S + G D N+ ++++Q+
Sbjct: 537 LVVSAAYDGTIKVWDLMKGYCTNTIMFPSICNALCFSTDGQTIFSGHVDGNLRLWDIQS- 713
Query: 181 QVEFKRIISPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLDD 223
R++S + + + + + + G +H+L D
Sbjct: 714 ----GRLLSEVATHSHAVTSISLSRNGNIALTSGGDNLHNLFD 830
>TC82717 weakly similar to GP|21539583|gb|AAM53344.1 putative coatomer
complex subunit {Arabidopsis thaliana}, partial (29%)
Length = 1118
Score = 43.1 bits (100), Expect = 2e-04
Identities = 35/129 (27%), Positives = 60/129 (46%), Gaps = 4/129 (3%)
Frame = +1
Query: 13 NKSFEVNQPPSDSVSSLCFSPK-ANLLVATSWDNQVRCWEVARNVATAPKASIS-HDQPV 70
+++FE N S V + F+PK + + S D ++ W + ++AP + H + +
Sbjct: 493 DETFEGN---SHYVMQVAFNPKDPSTFASASLDGTLKIWTID---SSAPNFTFEGHLKGM 654
Query: 71 LCSAW--KDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGS 128
C + +D + SG D K+W S T+ H V I PE+ +++T S
Sbjct: 655 NCVDYFESNDKQYLLSGSDDYTAKVWDYDSKNCVQTLEGHKNNVTAICAHPEIPIIITAS 834
Query: 129 WDRTIKYWD 137
D T+K WD
Sbjct: 835 EDSTVKIWD 861
Score = 33.1 bits (74), Expect = 0.16
Identities = 33/146 (22%), Positives = 59/146 (39%), Gaps = 11/146 (7%)
Frame = +1
Query: 78 DGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIP--EMSLLVTGSWDRTIKY 135
D +T S D +K+W + S T H + + + + L++GS D T K
Sbjct: 550 DPSTFASASLDGTLKIWTIDSSAPNFTFEGHLKGMNCVDYFESNDKQYLLSGSDDYTAKV 729
Query: 136 WDTRQPNPVHTQQLPERCYAMSVKH---PLMVVGTADRNI-----LVYNLQNP-QVEFKR 186
WD N V T + + H P+++ + D + + Y LQN +R
Sbjct: 730 WDYDSKNCVQTLEGHKNNVTAICAHPEIPIIITASEDSTVKIWDAVTYRLQNTLDFGLER 909
Query: 187 IISPLKYQTRCLAAFPDQQGFLVGSI 212
+ S + AF +GF++ ++
Sbjct: 910 VWSIGYKKGSSQLAFGCDKGFIIVTV 987
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.132 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,181,976
Number of Sequences: 36976
Number of extensions: 208360
Number of successful extensions: 1190
Number of sequences better than 10.0: 117
Number of HSP's better than 10.0 without gapping: 1049
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1147
length of query: 344
length of database: 9,014,727
effective HSP length: 97
effective length of query: 247
effective length of database: 5,428,055
effective search space: 1340729585
effective search space used: 1340729585
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0314.9


