

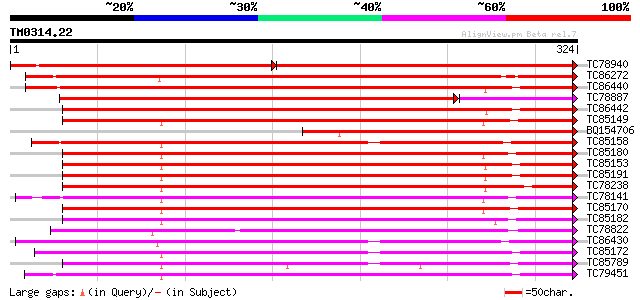
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0314.22
(324 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78940 similar to SP|P22196|PER2_ARAHY Cationic peroxidase 2 pr... 292 e-145
TC86272 similar to GP|4204761|gb|AAD11482.1| peroxidase precurso... 276 8e-75
TC86440 weakly similar to PIR|S14268|S14268 peroxidase (EC 1.11.... 273 7e-74
TC78887 similar to GP|5002236|gb|AAD37375.1| peroxidase {Glycine... 242 2e-73
TC86442 weakly similar to PIR|S14268|S14268 peroxidase (EC 1.11.... 257 4e-69
TC85149 similar to PIR|JC4780|JC4780 peroxidase (EC 1.11.1.7) 1B... 254 3e-68
BQ154706 similar to SP|P22196|PER2 Cationic peroxidase 2 precurs... 251 2e-67
TC85158 similar to SP|P22195|PER1_ARAHY Cationic peroxidase 1 pr... 251 3e-67
TC85180 homologue to PIR|JC4780|JC4780 peroxidase (EC 1.11.1.7) ... 249 1e-66
TC85153 similar to GP|2245683|gb|AAC98519.1| peroxidase precurso... 249 1e-66
TC85191 similar to PIR|JC4780|JC4780 peroxidase (EC 1.11.1.7) 1B... 248 2e-66
TC78238 similar to PIR|T05723|T05723 peroxidase (EC 1.11.1.7) pr... 247 4e-66
TC78141 similar to GP|5002348|gb|AAD37430.1| peroxidase 5 precur... 246 9e-66
TC85170 homologue to PIR|JC4781|JC4781 peroxidase (EC 1.11.1.7) ... 245 2e-65
TC85182 homologue to PIR|JC4780|JC4780 peroxidase (EC 1.11.1.7) ... 244 3e-65
TC78822 similar to PIR|T02962|T02962 peroxidase (EC 1.11.1.7) is... 242 1e-64
TC86430 similar to GP|23315187|gb|AAN20368.1 Sequence 378 from p... 242 2e-64
TC85172 weakly similar to GP|4760704|dbj|BAA77389.1 peroxidase 3... 242 2e-64
TC85789 similar to GP|5002344|gb|AAD37428.1| peroxidase 3 precur... 240 5e-64
TC79451 weakly similar to GP|4760700|dbj|BAA77387.1 peroxidase 1... 238 2e-63
>TC78940 similar to SP|P22196|PER2_ARAHY Cationic peroxidase 2 precursor (EC
1.11.1.7). [Peanut] {Arachis hypogaea}, partial (92%)
Length = 1272
Score = 292 bits (748), Expect(2) = e-145
Identities = 148/173 (85%), Positives = 154/173 (88%), Gaps = 1/173 (0%)
Frame = +3
Query: 153 SDVNNLPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTTACQFFSNRLYNFTSNGP- 211
SDVNNLPAP DSVD QKQKFA KGLNTQDLVTLVGGHTIGTTACQFFSNRL NFT+NG
Sbjct: 609 SDVNNLPAPGDSVDEQKQKFATKGLNTQDLVTLVGGHTIGTTACQFFSNRLRNFTTNGAA 788
Query: 212 DSSIDASFLPQLQALCPQNSGVSNRVALDTTSQNRFDTSYYANLRNGRGILLSDQALWND 271
D SID SFL QLQ LCPQNSG +NR+ALDT SQN+FD SYYANLRNGRGIL SDQALWND
Sbjct: 789 DPSIDPSFLSQLQTLCPQNSGATNRIALDTGSQNKFDNSYYANLRNGRGILQSDQALWND 968
Query: 272 ASTKTFVRRYLGLRGLLGLKFNVEFGKSMVKMSNIELKTGSDSEIRKICSAFN 324
ASTKTFV+RYLGLRGLLGL FNVEFG SMVKMSNI +KTG D EIRKICSAFN
Sbjct: 969 ASTKTFVQRYLGLRGLLGLTFNVEFGNSMVKMSNIGVKTGVDGEIRKICSAFN 1127
Score = 240 bits (612), Expect(2) = e-145
Identities = 122/153 (79%), Positives = 136/153 (88%), Gaps = 1/153 (0%)
Frame = +2
Query: 1 MERSLFRIAFLLLALASIVNT-VHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAA 59
ME SLF++ FLLL SIVNT V+GQG+RVGFY TC +AESIV+S V SHV SD +LA
Sbjct: 152 MEGSLFKVVFLLLVF-SIVNTLVYGQGTRVGFYSSTCSQAESIVKSTVASHVNSDSSLAP 328
Query: 60 GLLRMHFHDCFVQGCDASVLIAGAGTERTAPPNLGLRGYEVIDDAKAKVEAACPGVVSCA 119
GLLRMHFHDCFVQGCDASVL+AG+GTE+TA PNLGLRG+EVI+DAK K+EAACPGVVSCA
Sbjct: 329 GLLRMHFHDCFVQGCDASVLVAGSGTEKTAFPNLGLRGFEVIEDAKTKLEAACPGVVSCA 508
Query: 120 DILALAARDSVVLSGGLSWQVPTGRRDGRVSQA 152
DI+ALAARDSVVLSGGLSWQVPTGR DGRVSQA
Sbjct: 509 DIVALAARDSVVLSGGLSWQVPTGRXDGRVSQA 607
>TC86272 similar to GP|4204761|gb|AAD11482.1| peroxidase precursor {Glycine
max}, partial (86%)
Length = 1350
Score = 276 bits (706), Expect = 8e-75
Identities = 152/320 (47%), Positives = 204/320 (63%), Gaps = 5/320 (1%)
Frame = +1
Query: 10 FLLLALASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDC 69
F +L L + + H Q +GFY ++CP+AE IV + V H+++ +LAA L+RMHFHDC
Sbjct: 193 FKVLILCILAASTHAQ-LELGFYTKSCPKAEQIVANFVHEHIRNAPSLAAALIRMHFHDC 369
Query: 70 FVQGCDASVLIAGAG--TERTAPPNLGLRGYEVIDDAKAKVEAACPGVVSCADILALAAR 127
FV+GCDASVL+ E+ APPNL +RG++ ID K+ VEA CPGVVSCADI+AL+AR
Sbjct: 370 FVRGCDASVLLNSTNQQAEKNAPPNLTVRGFDFIDRIKSLVEAECPGVVSCADIIALSAR 549
Query: 128 DSVVLSGGLSWQVPTGRRDGRVSQASDVN-NLPAPFDSVDVQKQKFAAKGLNTQDLVTLV 186
DS+ +GG W+VPTGRRDG VS + N N+PAPF + + FA +GL+ +DLV L
Sbjct: 550 DSIAATGGPYWKVPTGRRDGVVSNLLEANQNIPAPFSNFTTLQTLFANQGLDMKDLVLLS 729
Query: 187 GGHTIGTTACQFFSNRLYNFTSNG-PDSSIDASFLPQLQAL-CPQNSGVSNRVALDTTSQ 244
G HTIG + C FSNRLYNFT G D S+D+ + L+ C + + V LD S+
Sbjct: 730 GAHTIGISLCTSFSNRLYNFTGKGDQDPSLDSEYAKNLKTFKCKNINDNTTIVELDPGSR 909
Query: 245 NRFDTSYYANLRNGRGILLSDQALWNDASTKTFVRRYLGLRGLLGLKFNVEFGKSMVKMS 304
N FD YY+ + RG+ SD AL ++ TK V ++ L+G L F EF KS+ KM
Sbjct: 910 NTFDLGYYSQVVKRRGLFESDSALLTNSVTKALVTQF--LQGSLE-NFYAEFAKSIEKMG 1080
Query: 305 NIELKTGSDSEIRKICSAFN 324
I++KTGS IRK C+ N
Sbjct: 1081QIKVKTGSQGVIRKHCALVN 1140
>TC86440 weakly similar to PIR|S14268|S14268 peroxidase (EC 1.11.1.7)
neutral - horseradish, partial (87%)
Length = 1272
Score = 273 bits (698), Expect = 7e-74
Identities = 154/323 (47%), Positives = 200/323 (61%), Gaps = 8/323 (2%)
Frame = +2
Query: 10 FLLLALASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDC 69
F+L+ + + TV Q + FY+ +CP IVR V +K++ + A LLR+HFHDC
Sbjct: 116 FVLINMFMLHFTVRSQLT-TDFYKSSCPNLTKIVRKEVVKALKNEMRMGASLLRLHFHDC 292
Query: 70 FVQGCDASVLIAGAGT-ERTAPPNLG-LRGYEVIDDAKAKVEAACPGVVSCADILALAAR 127
FV GCD S+L+ G E++A PN+ +RG++V+D K VE+AC GVVSCADILA+AAR
Sbjct: 293 FVNGCDGSILLDGGDDFEKSAFPNINSVRGFDVVDTIKTAVESACSGVVSCADILAIAAR 472
Query: 128 DSVVLSGGLSWQVPTGRRDGRVSQASDVN-NLPAPFDSVDVQKQKFAAKGLNTQDLVTLV 186
DSV+LSGG SW V GRRDG +S S N LP+PFD +D KF GLN D+V+L
Sbjct: 473 DSVLLSGGPSWSVMLGRRDGTISNGSLANVVLPSPFDPLDTIVSKFTNVGLNLTDVVSLS 652
Query: 187 GGHTIGTTACQFFSNRLYNFTSNG-PDSSIDASFLPQLQALCPQNSGVSNRVALDTTSQN 245
G HTIG C FSNRL+NF+ G PDS+++ L LQ LCPQ + LD S +
Sbjct: 653 GAHTIGRARCALFSNRLFNFSGTGSPDSTLETGMLTDLQNLCPQTGDGNTTAVLDRNSTD 832
Query: 246 RFDTSYYANLRNGRGILLSDQALWN----DASTKTFVRRYLGLRGLLGLKFNVEFGKSMV 301
FD YY NL NG+G+L SDQ L + ++++K V+ Y L F +F KSM+
Sbjct: 833 LFDNHYYKNLLNGKGLLSSDQILISTDEANSTSKPLVQSYNDNATL----FFGDFVKSMI 1000
Query: 302 KMSNIELKTGSDSEIRKICSAFN 324
KM NI KTGSD EIRK C N
Sbjct: 1001KMGNINPKTGSDGEIRKSCRVIN 1069
>TC78887 similar to GP|5002236|gb|AAD37375.1| peroxidase {Glycine max},
partial (75%)
Length = 1053
Score = 242 bits (618), Expect(2) = 2e-73
Identities = 124/229 (54%), Positives = 159/229 (69%), Gaps = 1/229 (0%)
Frame = +3
Query: 29 VGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQGCDASVLI-AGAGTER 87
VGFY TCP+ ES V V V DRT AA LLR+HFHDCFV+GCD S+LI E+
Sbjct: 87 VGFYSNTCPQVESTVHDVVREAVLFDRTKAAVLLRLHFHDCFVEGCDGSILINTTQNPEK 266
Query: 88 TAPPNLGLRGYEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPTGRRDG 147
TA P+ G++G+EVI+ AKA++EA+CPGVVSCADI+ALAARD++V++ G ++QVPTGRRDG
Sbjct: 267 TAFPHAGVKGFEVIERAKAQLEASCPGVVSCADIVALAARDAIVMANGPAYQVPTGRRDG 446
Query: 148 RVSQASDVNNLPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTTACQFFSNRLYNFT 207
VS S N+P DS+ K KF KGL +DLV L HTIGTTAC F RLY F
Sbjct: 447 FVSDKSLAGNMPDVNDSIQQLKTKFLNKGLTEKDLVLLSAAHTIGTTACFFMRKRLYEFF 626
Query: 208 SNGPDSSIDASFLPQLQALCPQNSGVSNRVALDTTSQNRFDTSYYANLR 256
G D +I+ +FLP+L+A CP++ V+ R+A+D S +FD S N+R
Sbjct: 627 PFGSDPTINLNFLPELKARCPKDGDVNIRLAMDEGSDLKFDKSILKNIR 773
Score = 51.6 bits (122), Expect(2) = 2e-73
Identities = 29/68 (42%), Positives = 39/68 (56%), Gaps = 1/68 (1%)
Frame = +1
Query: 258 GRGILLSDQALWNDASTKTFVRRYLG-LRGLLGLKFNVEFGKSMVKMSNIELKTGSDSEI 316
G +L SD L +D TK+ + Y + G F +F +SMVKM I +KTGS I
Sbjct: 778 GFAVLASDARLNDDFVTKSVIDSYFNPINPTFGPSFENDFVQSMVKMGQIGVKTGSVGNI 957
Query: 317 RKICSAFN 324
R++CSAFN
Sbjct: 958 RRVCSAFN 981
>TC86442 weakly similar to PIR|S14268|S14268 peroxidase (EC 1.11.1.7)
neutral - horseradish, partial (70%)
Length = 1151
Score = 257 bits (657), Expect = 4e-69
Identities = 144/303 (47%), Positives = 192/303 (62%), Gaps = 9/303 (2%)
Frame = +1
Query: 31 FYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQGCDASVLIAG-AGTERTA 89
F + CP +IVR V + + + +AA LLR+HFHDCFV GCDAS+L+ G E+ A
Sbjct: 97 FTGQACPDVFTIVRREVLNAINEEIRMAASLLRLHFHDCFVNGCDASILLDGDEDIEKFA 276
Query: 90 PPNLG-LRGYEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPTGRRDGR 148
PN+ RG+EVID K+ VE++C GVVSCADILA+ ARDSV LSGG W V GRRDG
Sbjct: 277 TPNINSARGFEVIDRIKSSVESSCSGVVSCADILAIVARDSVHLSGGPFWYVQLGRRDGL 456
Query: 149 VSQASDVNN-LPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTTACQFFSNRLYNFT 207
VS + NN +P+PFDS+D KF GL+ +D+VTL G HTIG C FFSNRL+NF+
Sbjct: 457 VSNKTLANNAIPSPFDSLDTIISKFDNVGLSVKDVVTLSGAHTIGRARCTFFSNRLFNFS 636
Query: 208 -SNGPDSSIDASFLPQLQALCPQNSGVSNRVALDTTSQNRFDTSYYANLRNGRGILLSDQ 266
+ PD+S++ L +LQ LCPQ+ + LD S ++FD +Y+ NL NG+G+L SDQ
Sbjct: 637 GTQEPDNSLEYEMLTELQNLCPQDGDGNTTTVLDPYSFDQFDNNYFKNLLNGKGLLSSDQ 816
Query: 267 ALWND-----ASTKTFVRRYLGLRGLLGLKFNVEFGKSMVKMSNIELKTGSDSEIRKICS 321
L++ ++TK V+ Y + F +EF +M+KM NI GS+ EIRK C
Sbjct: 817 ILFSSDEETTSTTKQLVQYYSENERI----FFMEFAYAMIKMGNINPLIGSEGEIRKSCR 984
Query: 322 AFN 324
N
Sbjct: 985 VIN 993
>TC85149 similar to PIR|JC4780|JC4780 peroxidase (EC 1.11.1.7) 1B precursor
- alfalfa, partial (95%)
Length = 1274
Score = 254 bits (649), Expect = 3e-68
Identities = 137/302 (45%), Positives = 192/302 (63%), Gaps = 8/302 (2%)
Frame = +2
Query: 31 FYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQGCDASVLIAGAGT---ER 87
FYR TCP+ SI+R + + K+D + A L+R+HFHDCFV GCDASVL+ T E+
Sbjct: 116 FYRDTCPKVHSIIREVIRNVSKTDPRMLASLVRLHFHDCFVLGCDASVLLNKTDTIVSEQ 295
Query: 88 TAPPNLG-LRGYEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPTGRRD 146
A PN+ LRG +V++ K VE ACP VSCADILAL+A+ S +L+ G +W+VP GRRD
Sbjct: 296 EAFPNINSLRGLDVVNQIKTAVEKACPNTVSCADILALSAQISSILADGPNWKVPLGRRD 475
Query: 147 GRVSQASDVN-NLPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTTACQFFSNRLYN 205
G + S N NLPAPF+S+D K FAA+GL+T DLV L G HT G C F ++RLYN
Sbjct: 476 GLTANQSLANQNLPAPFNSLDQLKSAFAAQGLSTTDLVALSGAHTFGRARCTFITDRLYN 655
Query: 206 FTSNG-PDSSIDASFLPQLQALCPQNSGVSNRVALDTTSQNRFDTSYYANLRNGRGILLS 264
F+S G PD +++ ++L +L+ +CP +N D T+ ++FD +YY+NL+ +G+L S
Sbjct: 656 FSSTGKPDPTLNTTYLQELRKICPNGGPPNNLANFDPTTPDKFDKNYYSNLQGKKGLLQS 835
Query: 265 DQALW--NDASTKTFVRRYLGLRGLLGLKFNVEFGKSMVKMSNIELKTGSDSEIRKICSA 322
DQ L+ + A T + V ++ + F F +M+KM NI + TG EIRK C+
Sbjct: 836 DQELFSTSGADTISIVNKFSADKN----AFFDSFEAAMIKMGNIGVLTGKKGEIRKHCNF 1003
Query: 323 FN 324
N
Sbjct: 1004VN 1009
>BQ154706 similar to SP|P22196|PER2 Cationic peroxidase 2 precursor (EC
1.11.1.7). [Peanut] {Arachis hypogaea}, partial (47%)
Length = 654
Score = 251 bits (642), Expect = 2e-67
Identities = 135/190 (71%), Positives = 141/190 (74%), Gaps = 33/190 (17%)
Frame = +2
Query: 168 QKQKFAAKGLNTQDLVTLVG--------------------------------GHTIGTTA 195
QKQKFA KGLNTQDLVTLVG GHTIGTTA
Sbjct: 2 QKQKFATKGLNTQDLVTLVGKRSKYFRNLFITLELHIDVASH*LIFYEYIIGGHTIGTTA 181
Query: 196 CQFFSNRLYNFTSNGP-DSSIDASFLPQLQALCPQNSGVSNRVALDTTSQNRFDTSYYAN 254
CQFFSNRL NFT+NG D SID SFL QLQ LCPQNSG +NR+ALDT SQN+FD SYYAN
Sbjct: 182 CQFFSNRLRNFTTNGAADPSIDPSFLSQLQTLCPQNSGATNRIALDTGSQNKFDNSYYAN 361
Query: 255 LRNGRGILLSDQALWNDASTKTFVRRYLGLRGLLGLKFNVEFGKSMVKMSNIELKTGSDS 314
LRNGRGIL SDQALWNDASTKTFV+RYLGLRGLLGL FNVEFG SMVKMSNI +KTG D
Sbjct: 362 LRNGRGILQSDQALWNDASTKTFVQRYLGLRGLLGLTFNVEFGNSMVKMSNIGVKTGVDG 541
Query: 315 EIRKICSAFN 324
EIRKICSAFN
Sbjct: 542 EIRKICSAFN 571
>TC85158 similar to SP|P22195|PER1_ARAHY Cationic peroxidase 1 precursor (EC
1.11.1.7). [Peanut] {Arachis hypogaea}, partial (86%)
Length = 1161
Score = 251 bits (641), Expect = 3e-67
Identities = 140/317 (44%), Positives = 192/317 (60%), Gaps = 5/317 (1%)
Frame = +2
Query: 13 LALASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQ 72
+ L ++ T+ Q S FY RTCP S ++ V S + ++R + A LLR+HFHDCFVQ
Sbjct: 104 IVLFCLIGTISAQLSS-NFYFRTCPLVLSTIKKEVISALINERRMGASLLRLHFHDCFVQ 280
Query: 73 GCDASVLIAGAGT---ERTAPPNLG-LRGYEVIDDAKAKVEAACPGVVSCADILALAARD 128
GCDASVL+ + E+TA PN LRG++VID K++VE CP VSCADILA+AARD
Sbjct: 281 GCDASVLLDDTSSFRGEKTAGPNANSLRGFDVIDKIKSEVEKLCPNTVSCADILAVAARD 460
Query: 129 SVVLSGGLSWQVPTGRRDGRVSQASDVN-NLPAPFDSVDVQKQKFAAKGLNTQDLVTLVG 187
SVV GGLSW V GRRD + N +LP P + F KG +++V L G
Sbjct: 461 SVVALGGLSWTVQLGRRDSTTASFGLANSDLPGPGSDLSGLINAFNNKGFTPKEMVALSG 640
Query: 188 GHTIGTTACQFFSNRLYNFTSNGPDSSIDASFLPQLQALCPQNSGVSNRVALDTTSQNRF 247
HTIG +C+FF R+YN +++ID+SF LQ+ CP+ G N LDTTS N F
Sbjct: 641 SHTIGEASCRFFRTRIYN------ENNIDSSFANSLQSSCPRTGGDLNLSPLDTTSPNTF 802
Query: 248 DTSYYANLRNGRGILLSDQALWNDASTKTFVRRYLGLRGLLGLKFNVEFGKSMVKMSNIE 307
D +Y+ NL+N +G+ SDQ L+++ +TK+ V Y+ L F V+F +M KM+N+
Sbjct: 803 DNAYFKNLQNQKGLFHSDQVLFDEVTTKSQVNSYV----RNPLSFKVDFANAMFKMANLG 970
Query: 308 LKTGSDSEIRKICSAFN 324
TGS ++RK C + N
Sbjct: 971 PLTGSSGQVRKNCRSVN 1021
>TC85180 homologue to PIR|JC4780|JC4780 peroxidase (EC 1.11.1.7) 1B
precursor - alfalfa, complete
Length = 1342
Score = 249 bits (636), Expect = 1e-66
Identities = 139/302 (46%), Positives = 188/302 (62%), Gaps = 8/302 (2%)
Frame = +1
Query: 31 FYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQGCDASVLIAGAGT---ER 87
FY +TCP SIVR + + K+D + A L+R+HFHDCFVQGCDASVL+ T E+
Sbjct: 109 FYSKTCPNVSSIVREVIRNVSKTDTRMLASLVRLHFHDCFVQGCDASVLLNNTATIVSEQ 288
Query: 88 TAPPNL-GLRGYEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPTGRRD 146
A PN LRG +V++ K VE ACP VSCADILALAA S LS G W+VP GRRD
Sbjct: 289 DAFPNRNSLRGLDVVNQIKTAVEKACPNTVSCADILALAAELSSTLSQGPDWKVPLGRRD 468
Query: 147 GRVSQASDVN-NLPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTTACQFFSNRLYN 205
G + S N NLPAPF+S+D K FA++GL+T DLV L G HT G C F +RLYN
Sbjct: 469 GLTANQSLANQNLPAPFNSLDQLKAAFASQGLSTTDLVALSGAHTFGRAHCSLFVSRLYN 648
Query: 206 FTSNG-PDSSIDASFLPQLQALCPQNSGVSNRVALDTTSQNRFDTSYYANLRNGRGILLS 264
F++ G PD +++A++L QL+ +CP + + D T+ ++FD +YY+NL+ +G+L S
Sbjct: 649 FSNTGSPDPTLNATYLQQLRNICPNGGPGTPLASFDPTTPDKFDKNYYSNLQVKKGLLQS 828
Query: 265 DQALW--NDASTKTFVRRYLGLRGLLGLKFNVEFGKSMVKMSNIELKTGSDSEIRKICSA 322
DQ L+ + A T + V + + F F +M+KM NI + TG+ EIRK C+
Sbjct: 829 DQELFSTSGADTISIVNNFATDQ----KAFFESFKAAMIKMGNIGVLTGNQGEIRKQCNF 996
Query: 323 FN 324
N
Sbjct: 997 VN 1002
>TC85153 similar to GP|2245683|gb|AAC98519.1| peroxidase precursor {Glycine
max}, complete
Length = 1413
Score = 249 bits (636), Expect = 1e-66
Identities = 140/302 (46%), Positives = 184/302 (60%), Gaps = 8/302 (2%)
Frame = +3
Query: 31 FYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQGCDASVLIAGAGT---ER 87
FY +TCP+ SI + K+D + A ++R+HFHDCFVQGCDASVL+ T E+
Sbjct: 141 FYGKTCPKLHSIAFKVLRKVAKTDPRMPASIIRLHFHDCFVQGCDASVLLNNTATIVSEQ 320
Query: 88 TAPPNLG-LRGYEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPTGRRD 146
A PN+ LRG +VI+ K KVE ACP VSCADIL LA+ S VL+GG W+VP GRRD
Sbjct: 321 DAFPNINSLRGLDVINQIKTKVEKACPNRVSCADILTLASGISSVLTGGPGWEVPLGRRD 500
Query: 147 GRVSQASDVN-NLPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTTACQFFSNRLYN 205
+ S N NLP P S+D K FAA+GLNT DLV L G HT G C F +RLYN
Sbjct: 501 SLTANQSLANQNLPGPNFSLDRLKSAFAAQGLNTVDLVALSGAHTFGRARCLFILDRLYN 680
Query: 206 FTSNG-PDSSIDASFLPQLQALCPQNSGVSNRVALDTTSQNRFDTSYYANLRNGRGILLS 264
F + G PD ++D ++L QL+ CPQN +NRV D T+ + D ++Y NL+ +G+L S
Sbjct: 681 FNNTGKPDPTLDTTYLQQLRNQCPQNGTGNNRVNFDPTTPDTLDKNFYNNLQGKKGLLQS 860
Query: 265 DQALWN--DASTKTFVRRYLGLRGLLGLKFNVEFGKSMVKMSNIELKTGSDSEIRKICSA 322
DQ L++ A T + V + + + F F SM+KM NI++ TG EIRK C+
Sbjct: 861 DQELFSTPGADTISIVNSFANSQNV----FFQNFINSMIKMGNIDVLTGKKGEIRKQCNF 1028
Query: 323 FN 324
N
Sbjct: 1029IN 1034
>TC85191 similar to PIR|JC4780|JC4780 peroxidase (EC 1.11.1.7) 1B precursor
- alfalfa, partial (96%)
Length = 1307
Score = 248 bits (634), Expect = 2e-66
Identities = 142/302 (47%), Positives = 185/302 (61%), Gaps = 8/302 (2%)
Frame = +2
Query: 31 FYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQGCDASVLIAGAGT---ER 87
FY +TCP+ +SIV +E K+D + A ++R+HFHDCFVQGCDASVL+ T E+
Sbjct: 179 FYAKTCPQLQSIVFQILEKVSKTDSRMPASIIRLHFHDCFVQGCDASVLLNKTSTIASEQ 358
Query: 88 TAPPNL-GLRGYEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPTGRRD 146
A PN+ LR +VI+ K +VE CP VSCADIL LAA S VLSGG W VP GRRD
Sbjct: 359 DAGPNINSLRRLDVINQIKTEVEKVCPNKVSCADILTLAAGVSSVLSGGPGWIVPLGRRD 538
Query: 147 GRVSQASDVN-NLPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTTACQFFSNRLYN 205
+ S N NLP P S+D K FAA+GLNT DLV L G HT+G C F +RLY+
Sbjct: 539 SLTANQSLANRNLPGPSSSLDQLKSSFAAQGLNTVDLVALSGAHTLGRARCLFILDRLYD 718
Query: 206 FTSNG-PDSSIDASFLPQLQALCPQNSGVSNRVALDTTSQNRFDTSYYANLRNGRGILLS 264
F + G PD ++D ++L QLQ CPQN +N V D T+ ++FD +YY NL+ +G+L S
Sbjct: 719 FDNTGKPDPTLDPTYLKQLQKQCPQNGPGNNVVNFDPTTPDKFDKNYYNNLQGKKGLLQS 898
Query: 265 DQALWN--DASTKTFVRRYLGLRGLLGLKFNVEFGKSMVKMSNIELKTGSDSEIRKICSA 322
DQ L++ A T + V + + + F F SM+KM NI + TG EIRK C+
Sbjct: 899 DQELFSTPGADTISIVNNFGNNQNV----FFQNFINSMIKMGNIGVLTGKKGEIRKQCNF 1066
Query: 323 FN 324
N
Sbjct: 1067VN 1072
>TC78238 similar to PIR|T05723|T05723 peroxidase (EC 1.11.1.7) precursor
seed coat - soybean, partial (90%)
Length = 1243
Score = 247 bits (631), Expect = 4e-66
Identities = 137/302 (45%), Positives = 184/302 (60%), Gaps = 8/302 (2%)
Frame = +2
Query: 31 FYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQGCDASVLIAGAGT---ER 87
FY +TCP IV + +D + A L+R+HFHDCFVQGCD SVL+ T E+
Sbjct: 107 FYSQTCPFLYPIVFRVIFEASLTDPRIGASLIRLHFHDCFVQGCDGSVLLNNTNTIVSEQ 286
Query: 88 TAPPNLG-LRGYEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPTGRRD 146
A PN+ LRG +V++ K VE CP VSCADIL +AA+ + VL GG SWQ+P GRRD
Sbjct: 287 DALPNINSLRGLDVVNQIKTAVENECPATVSCADILTIAAQVASVLGGGPSWQIPLGRRD 466
Query: 147 G-RVSQASDVNNLPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTTACQFFSNRLYN 205
+QA NLPAPF ++D K F +GLNT DLVTL G HT G C F NRLYN
Sbjct: 467 SLTANQALANQNLPAPFFTLDQLKAAFLVQGLNTTDLVTLSGAHTFGRAKCSTFINRLYN 646
Query: 206 FTSNG-PDSSIDASFLPQLQALCPQNSGVSNRVALDTTSQNRFDTSYYANLRNGRGILLS 264
F S G PD +++ ++L L+ +CPQN +N LD T+ N+FD +Y+NL++ +G+L S
Sbjct: 647 FNSTGNPDQTLNTTYLQTLREICPQNGTGNNLTNLDLTTPNQFDNKFYSNLQSHKGLLQS 826
Query: 265 DQALWN--DASTKTFVRRYLGLRGLLGLKFNVEFGKSMVKMSNIELKTGSDSEIRKICSA 322
DQ L++ +A T V + + L F V SM+KM+NI + TG++ EIR C+
Sbjct: 827 DQELFSTPNADTIAIVNSFSSNQALFFENFRV----SMIKMANISVLTGNEGEIRLQCNF 994
Query: 323 FN 324
N
Sbjct: 995 IN 1000
>TC78141 similar to GP|5002348|gb|AAD37430.1| peroxidase 5 precursor
{Phaseolus vulgaris}, partial (92%)
Length = 1254
Score = 246 bits (628), Expect = 9e-66
Identities = 142/330 (43%), Positives = 190/330 (57%), Gaps = 9/330 (2%)
Frame = +2
Query: 4 SLFRIAFLLLALASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLR 63
S+F + LL +N H Q + FY TCP SIVR+ V+ +++D + A L R
Sbjct: 56 SIFTVLIFLL-----LNPSHAQLTST-FYSNTCPSVSSIVRNVVQQALQNDPRITASLTR 217
Query: 64 MHFHDCFVQGCDASVLIAGAGT-----ERTAPPNLGLRGYEVIDDAKAKVEAACPGVVSC 118
+HFHDCFV GCDAS+L+ G + P N RG++V+D K VE +CP VVSC
Sbjct: 218 LHFHDCFVNGCDASLLLDQGGNITLSEKNAVPNNNSARGFDVVDKIKTSVENSCPSVVSC 397
Query: 119 ADILALAARDSVVLSGGLSWQVPTGRRDGRVSQASDVN-NLPAPFDSVDVQKQKFAAKGL 177
ADILALAA SV LSGG SW V GRRDG ++ S N ++P P +S+ KFAA GL
Sbjct: 398 ADILALAAEASVSLSGGPSWNVLLGRRDGLIANQSGANTSIPNPTESLANVTAKFAAVGL 577
Query: 178 NTQDLVTLVGGHTIGTTACQFFSNRLYNFTSNG-PDSSIDASFLPQLQALCPQNSGVSNR 236
NT DLV L G HT G C+FF+ RL+NF+ G PD ++++++L LQ CPQN +
Sbjct: 578 NTSDLVALSGAHTFGRGQCRFFNQRLFNFSGTGKPDPTLNSTYLATLQQNCPQNGSGNTL 757
Query: 237 VALDTTSQNRFDTSYYANLRNGRGILLSDQALW--NDASTKTFVRRYLGLRGLLGLKFNV 294
LD +S N FD +Y+ NL +G+L +DQ L+ N A+T + V + + F
Sbjct: 758 NNLDPSSPNNFDNNYFKNLLKNQGLLQTDQELFSTNGAATISIVNNFASNQ----TAFFE 925
Query: 295 EFGKSMVKMSNIELKTGSDSEIRKICSAFN 324
F +SM+ M NI GS EIR C N
Sbjct: 926 AFVQSMINMGNISPLIGSQGEIRSDCKKVN 1015
>TC85170 homologue to PIR|JC4781|JC4781 peroxidase (EC 1.11.1.7) 1C
precursor - alfalfa, complete
Length = 1164
Score = 245 bits (625), Expect = 2e-65
Identities = 134/302 (44%), Positives = 187/302 (61%), Gaps = 8/302 (2%)
Frame = +1
Query: 31 FYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQGCDASVLIAGAGT----E 86
FY +TCP SIV + + + K+D+ + A L+R+HFHDCFV GCDASVL+ T +
Sbjct: 103 FYSKTCPTVSSIVSNVLTNVSKTDQRMLASLVRLHFHDCFVLGCDASVLLNNTATIVSEQ 282
Query: 87 RTAPPNLGLRGYEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPTGRRD 146
+ P N LRG +V++ K +E+ACP VSCADILALAA+ S VL+ G SW VP GRRD
Sbjct: 283 QAFPNNNSLRGLDVVNQIKTAIESACPNTVSCADILALAAQASSVLAQGPSWTVPLGRRD 462
Query: 147 GRVSQASDVN-NLPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTTACQFFSNRLYN 205
G + + N NLPAPF+++ K F A+GLNT DLV L G HT G C F RLYN
Sbjct: 463 GLTANRTLANQNLPAPFNTLVQLKAAFTAQGLNTTDLVALSGAHTFGRAHCAQFVGRLYN 642
Query: 206 FTSNG-PDSSIDASFLPQLQALCPQNSGVSNRVALDTTSQNRFDTSYYANLRNGRGILLS 264
F+S G PD +++ ++L QL+ +CP +N D T+ ++FD +YY+NL+ +G+L S
Sbjct: 643 FSSTGSPDPTLNTTYLQQLRTICPNGGPGTNLTNFDPTTPDKFDKNYYSNLQVKKGLLQS 822
Query: 265 DQALW--NDASTKTFVRRYLGLRGLLGLKFNVEFGKSMVKMSNIELKTGSDSEIRKICSA 322
DQ L+ + A T + V ++ + F F +M+KM NI + TG+ EIRK C+
Sbjct: 823 DQELFSTSGADTISIVNKFSTDQN----AFFESFKAAMIKMGNIGVLTGTKGEIRKQCNF 990
Query: 323 FN 324
N
Sbjct: 991 VN 996
>TC85182 homologue to PIR|JC4780|JC4780 peroxidase (EC 1.11.1.7) 1B
precursor - alfalfa, complete
Length = 1373
Score = 244 bits (623), Expect = 3e-65
Identities = 136/302 (45%), Positives = 181/302 (59%), Gaps = 8/302 (2%)
Frame = +3
Query: 31 FYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQGCDASVLIAGAGT---ER 87
FYR TCP SIVR + S K D + L+R+HFHDCFVQGCDASVL+ T E+
Sbjct: 240 FYRNTCPNVSSIVREVIRSVSKKDPRMLGSLVRLHFHDCFVQGCDASVLLNKTDTVVSEQ 419
Query: 88 TAPPNL-GLRGYEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPTGRRD 146
A PN LRG +V++ K VE ACP VSCADILAL+A S L+ G W+VP GRRD
Sbjct: 420 DAFPNRNSLRGLDVVNQIKTAVEKACPNTVSCADILALSAELSSTLADGPDWKVPLGRRD 599
Query: 147 GRVSQASDVN-NLPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTTACQFFSNRLYN 205
G + N NLPAPF++ D K FAA+GL+T DLV L G HT G C F +RLYN
Sbjct: 600 GLTANQLLANKNLPAPFNTTDQLKAAFAAQGLDTTDLVALSGAHTFGRAHCSLFVSRLYN 779
Query: 206 FTSNG-PDSSIDASFLPQLQALCPQNSGVSNRVALDTTSQNRFDTSYYANLRNGRGILLS 264
F G PD +++ ++L QL+ +CP +N D T+ ++FD +YY+NL+ +G+L S
Sbjct: 780 FNGTGSPDPTLNTTYLQQLRTICPNGGPGTNLTNFDPTTPDKFDKNYYSNLQVKKGLLQS 959
Query: 265 DQALWNDASTKT--FVRRYLGLRGLLGLKFNVEFGKSMVKMSNIELKTGSDSEIRKICSA 322
DQ L++ + + T V ++ + F F +M+KM NI + TG EIRK C+
Sbjct: 960 DQELFSTSGSDTISIVNKFATDQ----KAFFESFKAAMIKMGNIGVLTGKQGEIRKQCNF 1127
Query: 323 FN 324
N
Sbjct: 1128VN 1133
>TC78822 similar to PIR|T02962|T02962 peroxidase (EC 1.11.1.7) isozyme 40K
precursor cationic - common tobacco, partial (70%)
Length = 1181
Score = 242 bits (618), Expect = 1e-64
Identities = 135/306 (44%), Positives = 180/306 (58%), Gaps = 5/306 (1%)
Frame = +2
Query: 24 GQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQGCDASVLI--- 80
G R FY+++CP+AE IV++ HV S L A L+R+HFHDCFV+GCDASVL+
Sbjct: 104 GGSLRKNFYKKSCPQAEEIVKNITLQHVSSRPELPAKLIRLHFHDCFVRGCDASVLLEST 283
Query: 81 AGAGTERTAPPNLGLRGYEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQV 140
AG E+ A PNL L G++VI+D K +E CPG+VSCADIL LA RD+ +W+V
Sbjct: 284 AGNTAEKDAIPNLSLAGFDVIEDIKEALEEKCPGIVSCADILTLATRDA--FKNKPNWEV 457
Query: 141 PTGRRDGRVSQASD-VNNLPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTTACQFF 199
TGRRDG VS++ + + N+PAPF ++ +Q FA K L DLV L G HTIG C F
Sbjct: 458 LTGRRDGTVSRSIEALINIPAPFHNITQLRQIFANKKLTLHDLVVLSGAHTIGVGHCNLF 637
Query: 200 SNRLYNFTSNG-PDSSIDASFLPQLQALCPQNSGVSNRVALDTTSQNRFDTSYYANLRNG 258
SNRL+NFT G D S++ ++ L+ C S + V +D S FD YY L
Sbjct: 638 SNRLFNFTGKGDQDPSLNPTYANFLKTKCQGLSDTTTTVEMDPNSSTTFDNDYYPVLLQN 817
Query: 259 RGILLSDQALWNDASTKTFVRRYLGLRGLLGLKFNVEFGKSMVKMSNIELKTGSDSEIRK 318
+G+ SD AL ++ V + KF EF +SM +M IE+ TGS+ EIR+
Sbjct: 818 KGLFTSDAALLTTKQSRNIVNELVSQN-----KFFTEFSQSMKRMGAIEVLTGSNGEIRR 982
Query: 319 ICSAFN 324
CS N
Sbjct: 983 KCSVVN 1000
>TC86430 similar to GP|23315187|gb|AAN20368.1 Sequence 378 from patent US
6410718, partial (93%)
Length = 1290
Score = 242 bits (617), Expect = 2e-64
Identities = 142/326 (43%), Positives = 184/326 (55%), Gaps = 5/326 (1%)
Frame = +2
Query: 4 SLFRIAFLLLALASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLR 63
S + + F+L+A ++I + FY +CP SIV V + +K + + A LLR
Sbjct: 173 SYYFLLFVLVAASAISEADAKKKLSKDFYCSSCPELLSIVNQGVINAIKKETRIGASLLR 352
Query: 64 MHFHDCFVQGCDASVLIAGA----GTERTAPPNLGLRGYEVIDDAKAKVEAACPGVVSCA 119
+HFHDCFV GCDAS+L+ G + A N RG+ VIDD KA VE ACPGVVSCA
Sbjct: 353 LHFHDCFVNGCDASILLDDTSSFIGEKTAAANNNSARGFNVIDDIKANVEKACPGVVSCA 532
Query: 120 DILALAARDSVVLSGGLSWQVPTGRRDGRVSQASDVNN-LPAPFDSVDVQKQKFAAKGLN 178
DIL LAARDSVV GG SW V GRRD + SD NN +PAPF ++ K FA +GL+
Sbjct: 533 DILTLAARDSVVHLGGPSWNVGLGRRDSITASRSDANNSIPAPFLNLSALKTNFANQGLS 712
Query: 179 TQDLVTLVGGHTIGTTACQFFSNRLYNFTSNGPDSSIDASFLPQLQALCPQNSGVSNRVA 238
+DLV L G HTIG C F +YN DS++D+ F LQ CP++ +
Sbjct: 713 AKDLVALSGAHTIGLARCVQFRAHIYN------DSNVDSLFRKSLQNKCPRSGNDNVLEP 874
Query: 239 LDTTSQNRFDTSYYANLRNGRGILLSDQALWNDASTKTFVRRYLGLRGLLGLKFNVEFGK 298
LD + FD Y+ NL + +L SDQ L+N +ST VR+Y KF F K
Sbjct: 875 LDHQTPTHFDNLYFKNLLAKKALLHSDQELFNGSSTDNLVRKY----ATDNAKFFKAFAK 1042
Query: 299 SMVKMSNIELKTGSDSEIRKICSAFN 324
MVKMS+I+ TGS+ +IR C N
Sbjct: 1043GMVKMSSIKPLTGSNGQIRTNCRKIN 1120
>TC85172 weakly similar to GP|4760704|dbj|BAA77389.1 peroxidase 3
{Scutellaria baicalensis}, partial (88%)
Length = 1282
Score = 242 bits (617), Expect = 2e-64
Identities = 141/315 (44%), Positives = 183/315 (57%), Gaps = 5/315 (1%)
Frame = +1
Query: 15 LASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQGC 74
++S++ T + FY CPRA I+R + + V DR L A LLR+HFHDCFVQGC
Sbjct: 130 VSSLIPTPNYSELSSTFYGTRCPRALYIIRREIIAAVSRDRRLGASLLRLHFHDCFVQGC 309
Query: 75 DASVLIAGAGT---ERTAPPNLG-LRGYEVIDDAKAKVEAACPGVVSCADILALAARDSV 130
DASVL+ T E+ A PN LRG+E ID KAK+EA CP VVSCADILA+AARDSV
Sbjct: 310 DASVLLKDTPTFQGEQNARPNANSLRGFEFIDSLKAKIEAVCPNVVSCADILAVAARDSV 489
Query: 131 VLSGGLSWQVPTGRRDGRVSQASDVN-NLPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGH 189
GG W V GRRD + + N +LP+PF ++ F KG + ++V L G H
Sbjct: 490 ATLGGPIWGVRLGRRDSTTANFNAANSDLPSPFLNLSGLIAAFKKKGFSADEMVALSGAH 669
Query: 190 TIGTTACQFFSNRLYNFTSNGPDSSIDASFLPQLQALCPQNSGVSNRVALDTTSQNRFDT 249
TIG C F NR+YN +S+I+ + LQ CP+N G +N LD+T+ FD+
Sbjct: 670 TIGKAKCAVFKNRIYN------ESNINPYYRRSLQNTCPRNGGDNNLANLDSTTPAFFDS 831
Query: 250 SYYANLRNGRGILLSDQALWNDASTKTFVRRYLGLRGLLGLKFNVEFGKSMVKMSNIELK 309
+YY NL RG+L SDQ L+N ST V Y L F +F K+M+KM N+
Sbjct: 832 AYYRNLLFKRGLLHSDQELYNGGSTDYKVLAYARNPYL----FRFDFAKAMIKMGNLSPL 999
Query: 310 TGSDSEIRKICSAFN 324
TG+ +IRK CS N
Sbjct: 1000TGNQGQIRKYCSRVN 1044
>TC85789 similar to GP|5002344|gb|AAD37428.1| peroxidase 3 precursor
{Phaseolus vulgaris}, partial (92%)
Length = 1467
Score = 240 bits (613), Expect = 5e-64
Identities = 135/303 (44%), Positives = 181/303 (59%), Gaps = 9/303 (2%)
Frame = +2
Query: 31 FYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQGCDASVLIAGAGT---ER 87
FY + CP V S V S V + + LLR+HFHDCFV GCD SVL+ + E+
Sbjct: 236 FYAKKCPNVFKAVNSVVHSAVAREPRMGGSLLRLHFHDCFVNGCDGSVLLDDTPSNKGEK 415
Query: 88 TAPPNL-GLRGYEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPTGRRD 146
TA PN LRG+EVID K+KVE+ CPGVVSCADI+A+AARDSVV GG W+V GRRD
Sbjct: 416 TALPNKDSLRGFEVIDAIKSKVESVCPGVVSCADIVAIAARDSVVNLGGPFWKVKLGRRD 595
Query: 147 GRVSQASDVNN--LPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTTACQFFSNRLY 204
+ + +D N+ +P PF +++ +F A+GL+T+D+V L G HTIG C + +R+Y
Sbjct: 596 SKTASLNDANSGVIPPPFSTLNNLINRFKAQGLSTKDMVALSGAHTIGKARCTVYRDRIY 775
Query: 205 NFTSNGPDSSIDASFLPQLQALCPQNSGV---SNRVALDTTSQNRFDTSYYANLRNGRGI 261
N D++ID+ F Q CP+ SG +N LD + N FD YY NL N +G+
Sbjct: 776 N------DTNIDSLFAKSRQRNCPRKSGTIKDNNVAVLDFKTPNHFDNLYYKNLINKKGL 937
Query: 262 LLSDQALWNDASTKTFVRRYLGLRGLLGLKFNVEFGKSMVKMSNIELKTGSDSEIRKICS 321
L SDQ L+N ST + V+ Y + F +F +M+KM N + TGS+ EIRK C
Sbjct: 938 LHSDQELFNGGSTDSLVKSYSNNQN----AFESDFAIAMIKMGNNKPLTGSNGEIRKQCR 1105
Query: 322 AFN 324
N
Sbjct: 1106RAN 1114
>TC79451 weakly similar to GP|4760700|dbj|BAA77387.1 peroxidase 1
{Scutellaria baicalensis}, partial (93%)
Length = 1229
Score = 238 bits (607), Expect = 2e-63
Identities = 141/323 (43%), Positives = 189/323 (57%), Gaps = 7/323 (2%)
Frame = +2
Query: 9 AFLLLALASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHD 68
A ++L + + H Q VGFY +C AE IV+ V + +AAGL+RMHFHD
Sbjct: 68 AIIVLVIYFLNGNAHSQ-LEVGFYTYSCGMAEFIVKDEVRKSFNKNPGIAAGLVRMHFHD 244
Query: 69 CFVQGCDASVLIAGAGT---ERTAPPNL-GLRGYEVIDDAKAKVEAACPGVVSCADILAL 124
CF++GCDASVL+ + E+ +P N LRG+EVID+AKAK+E C G+VSCADI+A
Sbjct: 245 CFIRGCDASVLLDSTLSNIAEKDSPANKPSLRGFEVIDNAKAKLEEECKGIVSCADIVAF 424
Query: 125 AARDSVVLSGGLSWQVPTGRRDGRVSQASDV-NNLPAPFDSVDVQKQKFAAKGLNTQDLV 183
AARDSV L+GGL + VP GRRDG++S ASD LP P +V+ Q FA KGL ++V
Sbjct: 425 AARDSVELAGGLGYDVPAGRRDGKISLASDTRTELPPPTFNVNQLTQLFAKKGLTQDEMV 604
Query: 184 TLVGGHTIGTTACQFFSNRLYNFTSNG-PDSSIDASFLPQLQALCPQNSGVSNRVA-LDT 241
TL G HTIG + C FS RLYNF+S D S+D S+ L+ CPQ + N V +D
Sbjct: 605 TLSGAHTIGRSHCSAFSKRLYNFSSTSIQDPSLDPSYAALLKRQCPQGNTNQNLVVPMDP 784
Query: 242 TSQNRFDTSYYANLRNGRGILLSDQALWNDASTKTFVRRYLGLRGLLGLKFNVEFGKSMV 301
+S D YY ++ RG+ SDQ L + T V + L ++ +F +MV
Sbjct: 785 SSPGTADVGYYNDILANRGLFTSDQTLLTNTGTARKVHQNARNPYL----WSNKFADAMV 952
Query: 302 KMSNIELKTGSDSEIRKICSAFN 324
KM + + TG+ EIR C N
Sbjct: 953 KMGQVGVLTGNAGEIRTNCRVVN 1021
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.135 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,949,480
Number of Sequences: 36976
Number of extensions: 108151
Number of successful extensions: 868
Number of sequences better than 10.0: 150
Number of HSP's better than 10.0 without gapping: 615
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 629
length of query: 324
length of database: 9,014,727
effective HSP length: 96
effective length of query: 228
effective length of database: 5,465,031
effective search space: 1246027068
effective search space used: 1246027068
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0314.22


