

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
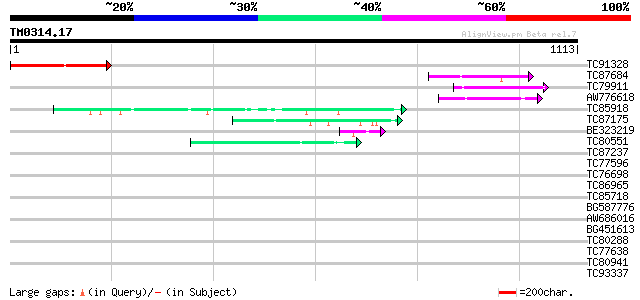
Query= TM0314.17
(1113 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC91328 187 2e-47
TC87684 similar to PIR|E85184|E85184 hypothetical protein dl4315... 145 6e-35
TC79911 similar to PIR|E85184|E85184 hypothetical protein dl4315... 119 7e-27
AW776618 homologue to GP|13359435|db putative senescence-associa... 94 3e-19
TC85918 similar to GP|13562014|gb|AAK30610.1 fibroin 1 {Plectreu... 47 5e-05
TC87175 similar to GP|21450874|gb|AAK59489.2 unknown protein {Ar... 45 1e-04
BE323219 45 2e-04
TC80551 similar to GP|17130993|dbj|BAB73602. ORF_ID:alr1903~unkn... 44 3e-04
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 41 0.003
TC77596 similar to GP|18377702|gb|AAL67001.1 putative clathrin b... 39 0.009
TC76698 similar to PIR|S50766|S50766 dehydrin-related protein - ... 38 0.019
TC86965 similar to GP|22022558|gb|AAM83236.1 At5g12410 {Arabidop... 35 0.12
TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa prot... 35 0.21
BG587776 weakly similar to PIR|F86242|F86 unknown protein 98896... 33 0.47
AW686016 weakly similar to GP|3581899|emb hypothetical serine-ri... 33 0.61
BG451613 33 0.61
TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein... 33 0.80
TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protei... 33 0.80
TC80941 weakly similar to PIR|T39903|T39903 serine-rich protein ... 33 0.80
TC93337 weakly similar to GP|21240660|gb|AAM44371.1 hypothetical... 32 1.0
>TC91328
Length = 675
Score = 187 bits (475), Expect = 2e-47
Identities = 111/211 (52%), Positives = 146/211 (68%), Gaps = 11/211 (5%)
Frame = +1
Query: 1 MADLFSRSYHCLWIPCRNVPS-KHSEQFRISAPFPFPFPRR--RISLTHHTHLVSNASSS 57
M++LFSRS+HCLW PC N+P K Q + P PRR + THH H++ +++SS
Sbjct: 10 MSNLFSRSFHCLWFPCSNLPPPKPPSQTLLPFSPSTPLPRRFFTFTTTHHPHILPSSTSS 189
Query: 58 SSSVP---DDVDILSSTECSDGSFVFQFAKASEIRDKLAELEREKLACE-VVEGGGKVGI 113
S+ P +++D++SSTE SDGSFVF F ASEIR+K+AEL ++KL E VVE + G+
Sbjct: 190 STPSPQEHEELDVISSTERSDGSFVFTFGNASEIREKIAELNKQKLVPEGVVE---EEGV 360
Query: 114 QALLS-DSIEILNNEVDDNLQEESKSSSIVVVGVADQNPQIPINEKE-SVVLDN--ESER 169
L+S D +EIL+NE D NL E +SSS +VV VADQNP + +E E SVVLD+ ESE
Sbjct: 361 SVLVSDDGVEILSNEDDSNLGGEIESSSTMVVDVADQNPLLLESENESSVVLDSEPESET 540
Query: 170 TVINESQKIDRHLKLDSIEDGDAQHGILSED 200
VIN+ Q ID+H KLDS+EDGD QHG+ SED
Sbjct: 541 PVINDLQVIDKHFKLDSVEDGDGQHGVCSED 633
>TC87684 similar to PIR|E85184|E85184 hypothetical protein dl4315c [imported]
- Arabidopsis thaliana, partial (65%)
Length = 1700
Score = 145 bits (367), Expect = 6e-35
Identities = 83/212 (39%), Positives = 125/212 (58%), Gaps = 5/212 (2%)
Frame = +2
Query: 822 RTELFLVSAAACLPHPSKALTGREDAYFISH-QNWVAVADGVGQWSPEGNNSGPYIRELM 880
R L ++S + LPHP K TG EDA+FI + + VADGVG W+ G N+ Y +EL+
Sbjct: 848 RKPLKMLSGSCYLPHPDKVATGGEDAHFICEDEQAIGVADGVGGWADVGVNARLYAQELV 1027
Query: 881 EKCKNIVSSHENISTIKPAEVLTRSAAETQTPGSSSVLVAYFDGEALHAANVGNTGFIII 940
C SS E + P VL ++ ++T+ GSS+V + EAL+A N+G++GFI+I
Sbjct: 1028 ANCAR-ASSEEPKGSFNPVRVLEKAHSKTKAMGSSTVCIIALIDEALNAINLGDSGFIVI 1204
Query: 941 RDGFIFKKSTAKFHEFNFPLHIVK----GDDPSEIIEGYKVDLCDGDVIIFATNGLFDNL 996
RDG + KS + FNFP + + GD PS E + V + GD+I+ T+GLFDN+
Sbjct: 1205 RDGSVIFKSPVQQRGFNFPYQLARSGTEGDLPSS-GEVFTVPVAPGDIIVAGTDGLFDNM 1381
Query: 997 YDQEIASTISKSLQASLGPQEIAEVLAKRAQE 1028
Y+ +I + + +A LGPQ A+ +A A++
Sbjct: 1382 YNNDIVGVVVGATRARLGPQATAQKIAALARQ 1477
>TC79911 similar to PIR|E85184|E85184 hypothetical protein dl4315c [imported]
- Arabidopsis thaliana, partial (57%)
Length = 924
Score = 119 bits (298), Expect = 7e-27
Identities = 69/190 (36%), Positives = 106/190 (55%), Gaps = 2/190 (1%)
Frame = +3
Query: 871 NSGPYIRELMEKCKNIVSSHENISTIKPAEVLTRSAAETQTPGSSSVLVAYFDGEALHAA 930
NSG Y RELM + + E ++ PA VL ++ + T+ GSS+ + + LHA
Sbjct: 15 NSGYYSRELMSNSVDAIRE-EPKGSVDPARVLEKAYSSTKAKGSSTACIIALTDQGLHAI 191
Query: 931 NVGNTGFIIIRDGFIFKKSTAKFHEFNFP--LHIVKGDDPSEIIEGYKVDLCDGDVIIFA 988
N+G++GF++IRDG +S + H+FNF L D + + V + GDVI+
Sbjct: 192 NLGDSGFMVIRDGCTIFQSPVQQHDFNFTYQLECSSHSDLPSSGQVFTVAVAPGDVIVAG 371
Query: 989 TNGLFDNLYDQEIASTISKSLQASLGPQEIAEVLAKRAQEVGKSTSTRSPFADAAQAIGY 1048
T+GLFDNLY+ EI + + ++A PQ A+ +A A++ ++PF+ AAQ G+
Sbjct: 372 TDGLFDNLYNNEITAVVVHGVRAGFSPQVTAQKIAALARQRALDKDRQTPFSTAAQDAGF 551
Query: 1049 VGYSGGKLDD 1058
Y GGKLDD
Sbjct: 552 -RYYGGKLDD 578
>AW776618 homologue to GP|13359435|db putative senescence-associated protein
{Pisum sativum}, partial (56%)
Length = 613
Score = 94.0 bits (232), Expect = 3e-19
Identities = 66/208 (31%), Positives = 107/208 (50%), Gaps = 5/208 (2%)
Frame = +3
Query: 843 GREDAYFISHQNW--VAVADGVGQWSPEGNNSGPYIRELMEKCKNIVSSHENISTIKPAE 900
G EDA+F+S+ N +AVADGV W+ E + RELM N V E + P
Sbjct: 6 GGEDAFFVSNYNGGVIAVADGVSGWAEEDVYPSLFPRELMANAYNFVQDEEVNND--PQI 179
Query: 901 VLTRSAAETQTPGSSSVLVAYFDGEA-LHAANVGNTGFIIIRDGFIFKKSTAKFHEFNFP 959
++ ++ A T + GS++V+VA + L ANVG+ G +IR+G + ++ + H F+ P
Sbjct: 180 LIRKAHAATFSTGSATVIVAMLEKNGNLKIANVGDCGLRVIRNGQVIFSTSPQEHYFDCP 359
Query: 960 LHIVKGDDPSEIIEGY--KVDLCDGDVIIFATNGLFDNLYDQEIASTISKSLQASLGPQE 1017
+ ++ V+L +GD I+ ++GLFDN++D EIA T+ A+ E
Sbjct: 360 YQLSSERVGQTYLDAMVSNVELMEGDTIVMGSDGLFDNVFDHEIALTV-----ANKEVSE 524
Query: 1018 IAEVLAKRAQEVGKSTSTRSPFADAAQA 1045
A+ L A + SP++ A+A
Sbjct: 525 AAQALTNLANSHSMDLNFDSPYSLEAKA 608
>TC85918 similar to GP|13562014|gb|AAK30610.1 fibroin 1 {Plectreurys tristis},
partial (8%)
Length = 4117
Score = 46.6 bits (109), Expect = 5e-05
Identities = 139/739 (18%), Positives = 276/739 (36%), Gaps = 47/739 (6%)
Frame = -1
Query: 87 EIRDKLAELEREKLACEVVEGGGKVGIQ---ALLSDSIEILNNEVDDNLQEESKSSSIVV 143
E ++K E E+ K A +VE + + A +S+ +I+ E + ++EE + +
Sbjct: 2578 ESQEKSEEAEQPK-AIAIVESTVEANEEKTAAAISEETKIIEVEPTETVKEEPMVTEVAT 2402
Query: 144 VGVADQNPQIPINE------KESVVLDNESERTVINESQ----KIDRHLKLDSI-EDGDA 192
+ + P + E +E V + E+ TV E K+ +K + + + D
Sbjct: 2401 AEIVKEEPVVTEVEATETLKEELVATEVEATETVKEEPAATEVKVTETVKDEPVVTEVDP 2222
Query: 193 QHGILSEDGAAECNGVPSVCEEE-----SQADDHKEVVVSRTVTAVSDVFSDINGGASEE 247
+ E A E +V EE + KE V+ V A V + A+ E
Sbjct: 2221 TETVKEEPLATEAEATETVKEEPVATEVEPTETVKEEPVATEVEATETVKDE---PAATE 2051
Query: 248 VEEKVDGQEEVVVSAVNPELDMFGDPVSSAFDGHKEVVS-AVTPELDVFGGLNSGASGEV 306
VE +EE V + + +PV++ + + V +V E++ + V
Sbjct: 2050 VEATETVKEEPVATETEATETVKEEPVATEVEATETVKEESVATEVEATETVKEEPVTTV 1871
Query: 307 EEEVDGLAVSAATQELDMFGDLNSGGALGEVEEKLDGDEDDAISTVTPESDMFSDLNNGA 366
E + + A E + + + EV+ + +++A++T E ++ +
Sbjct: 1870 VEPTEMVKDEPAATEFEATETVKDETIVTEVDPT-ETVKEEAVAT---EVEITKTVKEPL 1703
Query: 367 SGEVEEKLDGDEEDAISTVTP-----ESDMFSDMNNGASGEVEEKLDFHEADALSTDTPG 421
EV+ + ++ V P E + +++ A+ V+E+ E + T G
Sbjct: 1702 VTEVDLTESVKVKPVVTEVDPREMVKEEPVATEVQ--ATETVKEEPVATEVEPTET-VKG 1532
Query: 422 SNVNSDMNNGASGEVEEKID--DHEEASVSSITPESDMFTDISSGALGEVDEKLDVPEEV 479
V +++ + + + E+ + SSI +S D+ A+ E +L+
Sbjct: 1531 EPVVTEVEENQKEPEQHSTERREEEQPNASSIPEQSTETNDVI--AVEEKSRELEF---- 1370
Query: 480 AVSAVTPESDMFSDMNNGSAGEVEEKLDHEEVAVSTVTPEADMFTDVNSGALGEVDEKLD 539
E+ + + N AG E + V V E D E +
Sbjct: 1369 -------EAAILKETKNDEAGPAETE------KVEPVVTEVD-----------ENQSEPG 1262
Query: 540 VPEEVAVSAVTPESDMFSDMNNGSAGEVEEKLDHEEVATVST-------VTREADMFTDI 592
+ V P+ + + G EV D + A T + + + +
Sbjct: 1261 IQSFKQEEEVKPKEETENQEQTGDRVEVHPPKDSDIEAVKETNNSGSEAILVKEENIDPL 1082
Query: 593 NNGAPGEVEEKLDDKEEVSVSTVTLEADMFTDMNSSAPGEV-DEKLDGHEEV-------- 643
+NG ++ E+L EEV +E + T+ + G + +E+ + + V
Sbjct: 1081 SNGVEEKLSEQLQVGEEVGEIIKEVEPEQATEKTEGSNGTIKEEETEPNTTVNAAQVQTT 902
Query: 644 -ADLTSGASGEVEEKL---DALEEVVISIVTESDLFSDMNSGASEEVEEKEGRRTADKGH 699
A+L S EVEEK+ D +EVV++ E ++ E+ E + +
Sbjct: 901 PANLVE-PSPEVEEKIVVEDGKKEVVVAAEDEKKEPEASDAVQVSSREQNEAKTATIEAG 725
Query: 700 GVDVATSYLTGSVDADLSELVPASTLLDSEQAVNTEANNLTTSLESELVANDEETTHDIV 759
+ ++ +V+ + E + + D E+ + +NL E V + E+T
Sbjct: 724 ETETKVEEISKAVNEPVRETLASKFEKDEEETIQNGVDNLEKERIEEPVKTEVEST---- 557
Query: 760 DDSIDASEMEKSTMLHDLP 778
E + +T+ DLP
Sbjct: 556 ------KENDTTTISKDLP 518
>TC87175 similar to GP|21450874|gb|AAK59489.2 unknown protein {Arabidopsis
thaliana}, partial (10%)
Length = 1612
Score = 45.4 bits (106), Expect = 1e-04
Identities = 83/406 (20%), Positives = 144/406 (35%), Gaps = 72/406 (17%)
Frame = +3
Query: 438 EKIDDHEEASVSSITPESDMFTDISSGALGEVDEKLDVPEEVAVSAVTPESDMFSDMNN- 496
E + E V SI P DI+S AL + D +A SD D++N
Sbjct: 219 EDAEQFENGDVGSIKPAVVNGLDINSPALQDPHGSEDASSNLANKINNECSDASQDISNQ 398
Query: 497 GSAGEVEEKLDHEEVAVSTVTPEADMFTDVNSGALGEVDEKLDVPEEVAVSAVTPESDMF 556
G G H E +S E ++ + G + +K+ + ++ +P+ +
Sbjct: 399 GLTGRTVCSKSHSEERISNFVSEN--CASPSNSSHGVLKKKIKISMSSPINDPSPKCQLI 572
Query: 557 SDMNNGSAGEVEEKLDHEEVATVSTVTREADM------------------FTDINNGAPG 598
S + + G + D E+ + T +D+ FTD+N +PG
Sbjct: 573 SPNDTSTNGLPSDHSDSHEIRALGQATACSDLATTVKNMVEPASEGKPHAFTDLNCDSPG 752
Query: 599 EVEEKLDDKEEVSVSTVTLEADMFTD----------MNSSAPGEVDEKLDGHEEVADLTS 648
+ + D + V + E DM +NSS +E + + + +
Sbjct: 753 LNQNPVSDSQVVDNALSFKEFDMNDAHVEVLEAGPLVNSSQANNTEENNENVDVSGSVLN 932
Query: 649 GASGEVEEKLDALEE--VVISIVTESDLF---SDMNSGASEEVE------EKEGRRTADK 697
A ++ L++ V S V ++D+ DMN E VE +G +
Sbjct: 933 HAITDMNCDSPGLDQNPVSDSRVADNDMSLKKFDMNDAHVEVVEAGPLVNSFKGNNAKEN 1112
Query: 698 GHGVDVATSYLTGS---------------------VDADLS-----------ELVPASTL 725
VDV+ S L + VD DLS ++ A L
Sbjct: 1113DENVDVSGSVLNHAITDMNCDSPGLDQNPVSDSQVVDNDLSLKKFDMNDSLGLVLEAGPL 1292
Query: 726 LDSEQAVNTEANNLTTSLESELVANDEETTHDIVDDSIDASEMEKS 771
+S QA NT+ NN + + DI D + D+ ++++
Sbjct: 1293GNSSQANNTKENNENVDVSGSAL------NQDITDMNCDSPGLDQN 1412
>BE323219
Length = 603
Score = 44.7 bits (104), Expect = 2e-04
Identities = 33/99 (33%), Positives = 45/99 (45%), Gaps = 7/99 (7%)
Frame = +3
Query: 647 TSGASGEVEEKLDALEEVVISIVTESD-------LFSDMNSGASEEVEEKEGRRTADKGH 699
T +G V + LE+V S +E D L + S +E V + G
Sbjct: 132 TDNVTGSVCTDISELEQVSTSSESEQDGYIARDNLTDSVESDITESVPVSTFSESEQDGF 311
Query: 700 GVDVATSYLTGSVDADLSELVPASTLLDSEQAVNTEANN 738
AT LTGS DAD+SE +P ST ++SEQ E +N
Sbjct: 312 S---ATDNLTGSFDADMSESMPVSTTIESEQVGYIETDN 419
Score = 42.4 bits (98), Expect = 0.001
Identities = 20/40 (50%), Positives = 29/40 (72%)
Frame = +1
Query: 708 LTGSVDADLSELVPASTLLDSEQAVNTEANNLTTSLESEL 747
L GSVD ++S+LVP ST L+SEQ + +NLT S ++E+
Sbjct: 421 LIGSVDVEMSDLVPVSTTLESEQVGYSATDNLTGSADAEM 540
Score = 39.7 bits (91), Expect = 0.007
Identities = 27/92 (29%), Positives = 48/92 (51%), Gaps = 6/92 (6%)
Frame = +3
Query: 662 EEVVISIVTESD-----LFSDMNSGASEEVEEKEGRRTADKGHGVD-VATSYLTGSVDAD 715
E V +SI +ES+ ++ ++ E E T+ + +A LT SV++D
Sbjct: 78 ESVPVSISSESEQGGYGATDNVTGSVCTDISELEQVSTSSESEQDGYIARDNLTDSVESD 257
Query: 716 LSELVPASTLLDSEQAVNTEANNLTTSLESEL 747
++E VP ST +SEQ + +NLT S ++++
Sbjct: 258 ITESVPVSTFSESEQDGFSATDNLTGSFDADM 353
Score = 36.6 bits (83), Expect = 0.056
Identities = 30/101 (29%), Positives = 48/101 (46%), Gaps = 6/101 (5%)
Frame = +3
Query: 694 TADKGHGVDV-ATSYLTGSVDADLSELVPASTLLDSEQAVNTEANNLTTSL-----ESEL 747
T+ + VD A LTG +DA SE VP S +SEQ +N+T S+ E E
Sbjct: 3 TSSESEQVDCSARENLTGIIDAKTSESVPVSISSESEQGGYGATDNVTGSVCTDISELEQ 182
Query: 748 VANDEETTHDIVDDSIDASEMEKSTMLHDLPASSDMENKID 788
V+ E+ D + ++ +S + +P S+ E++ D
Sbjct: 183 VSTSSESEQDGYIARDNLTDSVESDITESVPVSTFSESEQD 305
>TC80551 similar to GP|17130993|dbj|BAB73602. ORF_ID:alr1903~unknown protein
{Nostoc sp. PCC 7120}, partial (4%)
Length = 1226
Score = 44.3 bits (103), Expect = 3e-04
Identities = 74/349 (21%), Positives = 123/349 (35%), Gaps = 14/349 (4%)
Frame = +3
Query: 356 SDMFSDLNNGASGEVEEK--LDGDEEDAISTVTPESDMFSDMNNGASGEVEEKLDFHEAD 413
S++ + NG S EK L G +E P S+ +NG S EK
Sbjct: 18 SEVMKNSENGVSSNHHEKETLAGLQEIKSEAAFP-SEAMQYYDNGVSTNHHEKETLAGLQ 194
Query: 414 ALSTDTP-GSNVNSDMNNGASGEVEEKIDDH--EEASVSSITPESDMFTDISSGALGEVD 470
+ ++ S + NGAS EK +E + P + + +G
Sbjct: 195 EVKSEAAFPSEAMKNSENGASTNYHEKETSAGLQEVKSEAAFPPDEAMQNSENGVPTNHH 374
Query: 471 EK--LDVPEEVAVSAVTPESDMFSDMNNGSAGEVEEK---LDHEEVAVSTVTPEADMFTD 525
EK L +EV A P + + NG++ EK +EV P + +
Sbjct: 375 EKETLGAIKEVKSEAAFPSDEAMQNSENGASTNHHEKETLAAIQEVKSEAAFPSDEAMQN 554
Query: 526 VNSGALGEVDEK--LDVPEEVAVSAVTPESDMFSDMNNGSAGEVEEKLDHEEVATVSTVT 583
+G EK L +E+ A P + + NG + EK E +A +
Sbjct: 555 SENGVSTNHHEKETLATIQEIKSEAAFPSDETMMNSENGVSTNHHEK---ETLAAIQENK 725
Query: 584 READMFTD--INNGAPGEVEEKLDDKEEVSVSTVTLEADMFTDMNSSAPGEVDEKLDGHE 641
EA +D N G + + ++ + EA ++ +++ V H
Sbjct: 726 SEAAFPSDEATMNSENGVSTNHHEKETLAAIQEIKSEAAFLSEAMTNSENRVSTN---HH 896
Query: 642 EVADLTSGASGEVEEKLDALEEVVISIVTESDLFSDMNSGASEEVEEKE 690
E EEKL ++E+ S++ + S S + EKE
Sbjct: 897 E------------EEKLAGVQEIKSEAAFLSEVMKNSGSDISTNLHEKE 1007
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 40.8 bits (94), Expect = 0.003
Identities = 116/599 (19%), Positives = 217/599 (35%), Gaps = 35/599 (5%)
Frame = +1
Query: 45 THHTHLVSNASSSSSSVPDDVDILSSTECSDGSFVFQFAKASEIRDKLAELEREKLACEV 104
+H + S + SS ++ S E + S + E +D+ E+ K E
Sbjct: 10 SHLENEESKETGESSEEKSHLENEESKETGESSEEKSHLENEENKDE----EKSKQENEE 177
Query: 105 VEGGGKVGIQALLSDSIEILNNEVDDNLQEE-SKSSSIVVVGVADQNPQIPINEKESVVL 163
++ G K+ + + E E ++N EE S+ + + + I E++S
Sbjct: 178 IKDGEKIQQENEENKDEEKSQQENEENKDEEKSQQENELKKNEGGEKETGEITEEKSKQE 357
Query: 164 DNESERTVINESQKIDRHLK-LDSIEDGDAQHGILSEDGAAECNGVPSVCEEE------- 215
+ E+ T + + + + D+ E H S+ G E NG +EE
Sbjct: 358 NEETSETNSKDKENEESNQNGSDAKEQVGENHEQDSKQGTEETNGTEGGEKEEHDKIKED 537
Query: 216 ----SQADDHKEVVVSR--------TVTAVSDVFSDINGGASEEVEEKVDGQEEVVVSAV 263
+Q D ++ +R +AV D S + +EE +K + E + S
Sbjct: 538 TSSDNQVQDGEKNNEAREENYSGDNASSAVVDNKSQESSNKTEEQFDKKEKNEFELESQK 717
Query: 264 NPE--LDMFGDPVSSAFDGHKEVVSAVTPELDVFGGLNSGASGEVEEEVDGLAVSAATQE 321
N + ++ G++ E D G S + + +E+ Q
Sbjct: 718 NSNETTESTDSTITQNSQGNESEKDQAQTENDTPKGSASESDEQKQEQEQNNTTKDDVQT 897
Query: 322 LDMFGDLNSGGALGEVEEKLDGDEDDAISTVTPESDMFSDLNNGASGEVEEKLDGDEEDA 381
D S + EK + +DA S S + + NN S + + GD+ D+
Sbjct: 898 TD-----TSSQNGNDTTEKQNETSEDANSKKEDSSALNTTPNNEDS---KSGVAGDQADS 1053
Query: 382 ISTVTPESDMFSDMNNGASGEVEEKLDFHEADALSTDTPGSNVNS-DMNNGASGEV---- 436
+T + + N+G + + + T GS+ N+ D + AS +V
Sbjct: 1054TTTTSSSETQDGNTNHGEYKDTTNENPEKNSGQEGTQESGSSSNTFDNKDAASNKVQLTT 1233
Query: 437 ------EEKIDDHEEASVSSITPESDMFTDISSGALGEVDEKLDVPEEVAVSAVTPESDM 490
E+K D+ A S + ++D + +SG ++ + S VT S+
Sbjct: 1234TSDTSSEQKKDESSSAESKSESSQND---NANSGQSNTTSDE-SANDNKDSSQVTTSSEN 1401
Query: 491 FSDMNNGSAGEVEEKLDHEEVAVSTVTPEADMFTDVNSGALGEVDEKLDVPEEVAVSAV- 549
++ N+ + +E + D + N+ G +V E +A
Sbjct: 1402SAEGNSNTENNSDEN-------------QNDSKNNENTNDSGNTSNDANVNENQNENAAQ 1542
Query: 550 TPESDMFSDMNNGSAGEVEEKLDHEEVATVSTVTREADMFTDINNGAPGEVEEKLDDKE 608
T S+ D N S VE K ++ E A ++ D ++ N+ + DDKE
Sbjct: 1543TKTSENEGDAQNES---VESKKENNESA-----HKDVDNNSNSNDQGSSDTSVTQDDKE 1695
>TC77596 similar to GP|18377702|gb|AAL67001.1 putative clathrin binding
protein {Arabidopsis thaliana}, partial (44%)
Length = 2019
Score = 39.3 bits (90), Expect = 0.009
Identities = 60/296 (20%), Positives = 115/296 (38%), Gaps = 18/296 (6%)
Frame = +2
Query: 356 SDMFSDLNNGASGEVEEKLDGDEEDAISTVTPESDMFSDMNNGASGEVEEKLDFHEADAL 415
SD + D ++++ G +S+ E+ S E K
Sbjct: 701 SDSYGDKGRYDEAKIDKDYSGKSRHGVSSKNEENSFKKGSVRSVSKSQENK--------- 853
Query: 416 STDTPGSNVNSDMNNGASGEVEEKIDDHEEASVSSITP--ESDMFTDISSGALGE-VDEK 472
S+ S+ N+ + AS E+ DD + SS T S+ LG+ +D
Sbjct: 854 SSRASKSSANAVPSQSASVPTEDDFDDFDPRGTSSKTSAGSSNQVDLFGQDLLGDFMDAP 1033
Query: 473 LDVPEEVAVSAVTPESDMFSDMN---------NGSAGEVEEKLD--HEEVAVSTVTPEAD 521
V E ++ + D+F+D + G++ + ++++D + A+ +VTP D
Sbjct: 1034TSVSVEKPATSNVSDVDLFADASFVSAAPHADKGASSQPQDEVDLFSSQPAIPSVTPTVD 1213
Query: 522 MFTDVNSGALGEVDEKLDVPEEVAVSAVTPESDMFSDMNNGSAGEVEEKLDHEEVATVST 581
+F+ + S A+ + D K + V S P + + +NN +V + + S
Sbjct: 1214LFS-IPSPAV-QPDSKSENSVPVNNSTFDPFASV--PLNNFEGSDVFGDFTSQSDSVSSQ 1381
Query: 582 VTREADMFTDINNGAPGEVEEKLDDKEEVSVSTVTLEADMFT----DMNSSAPGEV 633
+ A TD + + + K++ + AD + D+N SAP +V
Sbjct: 1382PSTNA--ATDGSTSGKSVTDSNVSPKKDAFQVKSGIWADSLSRGLIDLNISAPKKV 1543
>TC76698 similar to PIR|S50766|S50766 dehydrin-related protein - garden pea,
partial (80%)
Length = 974
Score = 38.1 bits (87), Expect = 0.019
Identities = 47/192 (24%), Positives = 79/192 (40%), Gaps = 26/192 (13%)
Frame = +2
Query: 211 VCEEESQADDHKEVVVSRTVTAVSD--VFSDINGGASEEVEEKVDGQEEVVVSAVNPELD 268
+ EE+Q + + T + D VF D GG ++ E K QEE + + N ++
Sbjct: 83 IMAEENQNKYEETTATNSETTEIKDRGVF-DFLGGKKKDEEHKP--QEEAIATDFNHKVT 253
Query: 269 MFGDPVSSAFDGHKEVVSAVTPELDVFGGLNSGASGEVEEEVDGLA-----------VSA 317
++ P + + +E T L+ +S +S EEEVDG S
Sbjct: 254 LYEAPSETKVEEKEEGEKKHTSLLEKLHRSDSSSSSSSEEEVDGEKRKKKKKEKKEDTSV 433
Query: 318 ATQELDMFGDLNSGGALGEVEEKLDGDE--DD-----------AISTVTPESDMFSDLNN 364
A +++D + G L +++EKL G + DD A + T S D
Sbjct: 434 AVEKVDGTTE-EKKGFLDKIKEKLPGHKKTDDVTTPPAPPVVVAPAETTTTSHDQGDQKK 610
Query: 365 GASGEVEEKLDG 376
G +++EK+ G
Sbjct: 611 GILEKIKEKIPG 646
Score = 29.3 bits (64), Expect = 8.9
Identities = 27/121 (22%), Positives = 47/121 (38%)
Frame = +2
Query: 569 EKLDHEEVATVSTVTREADMFTDINNGAPGEVEEKLDDKEEVSVSTVTLEADMFTDMNSS 628
EKL + ++ S+ E D GE +K +++ S + D T+
Sbjct: 326 EKLHRSDSSSSSSSEEEVD----------GEKRKKKKKEKKEDTSVAVEKVDGTTEEKKG 475
Query: 629 APGEVDEKLDGHEEVADLTSGASGEVEEKLDALEEVVISIVTESDLFSDMNSGASEEVEE 688
++ EKL GH++ D+T+ + V T S D G E+++E
Sbjct: 476 FLDKIKEKLPGHKKTDDVTTPPAPPV-------VVAPAETTTTSHDQGDQKKGILEKIKE 634
Query: 689 K 689
K
Sbjct: 635 K 637
>TC86965 similar to GP|22022558|gb|AAM83236.1 At5g12410 {Arabidopsis
thaliana}, partial (59%)
Length = 1455
Score = 35.4 bits (80), Expect = 0.12
Identities = 48/206 (23%), Positives = 82/206 (39%), Gaps = 21/206 (10%)
Frame = +1
Query: 249 EEKVDGQEEVVVSAVNPELDM---FGDPVSSAFDGHKEVVSAVT------------PELD 293
EE +DG V + L+ F D SS+ D +E P+LD
Sbjct: 247 EELIDGAHPSVTKLSDKPLNKKITFADSDSSSIDDDEEEEEKAQVPEGGEEKEDKKPKLD 426
Query: 294 VFGGLNSGASGEVEEE-----VDGLAVSAATQELDMFGDLNSGGALGEVEEKLDGDEDDA 348
V N+ E E+ +DGL A + D GD++S + ++ ++L + +
Sbjct: 427 VSNSDNTSHDNETGEKSDPPKIDGLHAQAGDKANDDKGDVDSPKTIEKIADELPAVK-EC 603
Query: 349 ISTVTPESDMFSDLNNGASGEVEEKLDGDEEDAISTVTPES-DMFSDMNNGASGEVEEKL 407
T P S N G E+ +D ED + + +S F+ + +G +G V ++
Sbjct: 604 CKTTAPTS------NLGEKKVEEKSIDKLIEDELVELRDKSKKRFAKLESGCNGVVFIQM 765
Query: 408 DFHEADALSTDTPGSNVNSDMNNGAS 433
+ D +P VN + + AS
Sbjct: 766 RKKDGD----KSPNKIVNRIVTSAAS 831
>TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa protein
mitochondrial precursor. [Kidney bean French bean]
{Phaseolus vulgaris}, complete
Length = 2371
Score = 34.7 bits (78), Expect = 0.21
Identities = 26/75 (34%), Positives = 34/75 (44%)
Frame = +2
Query: 442 DHEEASVSSITPESDMFTDISSGALGEVDEKLDVPEEVAVSAVTPESDMFSDMNNGSAGE 501
D E S+ I +D +L E EK +P EVA SD+ S M SA E
Sbjct: 1805 DQERKSLIDIRNSADTSIYSIEKSLSEYREK--IPAEVAKEIEDAVSDLRSAMAGDSADE 1978
Query: 502 VEEKLDHEEVAVSTV 516
++ KLD AVS +
Sbjct: 1979 IKSKLDAANKAVSKI 2023
Score = 31.6 bits (70), Expect = 1.8
Identities = 31/123 (25%), Positives = 52/123 (42%), Gaps = 3/123 (2%)
Frame = +2
Query: 453 PESDMFTDISSGALGEV---DEKLDVPEEVAVSAVTPESDMFSDMNNGSAGEVEEKLDHE 509
P+ ++ DI + + V D+ +++ + + SD N E+ + D E
Sbjct: 1637 PQIEVTFDIDANGIVTVSAKDKSTGKEQQITIKSSGGLSDD-EIQNMVKEAELHAQKDQE 1813
Query: 510 EVAVSTVTPEADMFTDVNSGALGEVDEKLDVPEEVAVSAVTPESDMFSDMNNGSAGEVEE 569
++ + AD +L E EK +P EVA SD+ S M SA E++
Sbjct: 1814 RKSLIDIRNSADTSIYSIEKSLSEYREK--IPAEVAKEIEDAVSDLRSAMAGDSADEIKS 1987
Query: 570 KLD 572
KLD
Sbjct: 1988 KLD 1996
>BG587776 weakly similar to PIR|F86242|F86 unknown protein 98896-95855
[imported] - Arabidopsis thaliana, partial (23%)
Length = 608
Score = 33.5 bits (75), Expect = 0.47
Identities = 43/165 (26%), Positives = 70/165 (42%), Gaps = 4/165 (2%)
Frame = +2
Query: 17 RNVPSKHSEQFRISAPFPFPFPRRRISLTHHTHLVSNASSSSSSVPDDVDILSSTECSDG 76
R PS + + R +P + R+R + + ++SSS SV I + +
Sbjct: 86 RRSPSTTTWRRRSRSPTAKRYRRQRSRSSSLSPAHKSSSSSLGSVEQKTAIEKQRKEEEK 265
Query: 77 SFVFQFAKASEIRDKLAELEREKLACEVVEGGG----KVGIQALLSDSIEILNNEVDDNL 132
Q A+ I ++ A+ E + V E +V IQ L + + LN+EV L
Sbjct: 266 KRRQQEAELKLIAEETAKRVEEAIRKRVDESLSSEEVRVEIQRRLEEGRKKLNDEVAAQL 445
Query: 133 QEESKSSSIVVVGVADQNPQIPINEKESVVLDNESERTVINESQK 177
Q+E +++ I A Q + EKE + E R I ESQ+
Sbjct: 446 QKEKEAALIE----AKQKEEQARKEKEDLERMLEENRRKIEESQR 568
>AW686016 weakly similar to GP|3581899|emb hypothetical serine-rich protein
{Schizosaccharomyces pombe}, partial (4%)
Length = 593
Score = 33.1 bits (74), Expect = 0.61
Identities = 31/125 (24%), Positives = 50/125 (39%), Gaps = 2/125 (1%)
Frame = +2
Query: 567 VEEKLDHEEVATVSTVTREADMFTDINNGAPGEVEEKLDDKEEVSVSTVTLEADMFTDMN 626
VE+ +D E +A E + D ++E + + E+ + TV E + +
Sbjct: 161 VEDSMDSERIAEEGDEECELERVEDFVEFVEDSLDEGIVEDEDYVLETV--ERNFGAEAE 334
Query: 627 SSAPGEVDEKLDGHEEVAD--LTSGASGEVEEKLDALEEVVISIVTESDLFSDMNSGASE 684
E+ D EEV + L G VEE+ + E V +++ SD A E
Sbjct: 335 KVRRNELQPGFDEDEEVRENELEPNFEGVVEEEDEVNVEAAAKEVGVNEIQSDFVEVAEE 514
Query: 685 EVEEK 689
EV K
Sbjct: 515 EVGRK 529
>BG451613
Length = 636
Score = 33.1 bits (74), Expect = 0.61
Identities = 27/108 (25%), Positives = 51/108 (47%), Gaps = 1/108 (0%)
Frame = +3
Query: 307 EEEVDGLAVSAATQELDMFGDLNSGGALGEVEEKLDGDED-DAISTVTPESDMFSDLNNG 365
+E GLA T ELD+ + G++ L+ D+D +T+ D DL N
Sbjct: 174 QEIAAGLAYDDKTSELDVQDESQEDRQDGKMTIPLNDDKDVQGKTTILASDDDPFDLKNS 353
Query: 366 ASGEVEEKLDGDEEDAISTVTPESDMFSDMNNGASGEVEEKLDFHEAD 413
+GE E+ + E D + + S+ ++++ + + ++ D H+AD
Sbjct: 354 FAGEEEDNI--SETDKKNLLEDTSEHANEISQEENQNIPQE-DAHDAD 488
>TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein T28I19.100
- Arabidopsis thaliana, partial (14%)
Length = 1460
Score = 32.7 bits (73), Expect = 0.80
Identities = 45/201 (22%), Positives = 66/201 (32%), Gaps = 5/201 (2%)
Frame = +3
Query: 565 GEVEEKLDHEEVATVSTVTREADMFTDINNGAPGEVEEKLDDKEEVSVSTVTLEADMFTD 624
GE K + EE + + R + + N EE EEV +E D
Sbjct: 477 GEEGNKHETEEESEDNVHERREEQDEEENKHGAEVQEENESKSEEVEDEGGDVEIDENDH 656
Query: 625 MNSSAPGE-----VDEKLDGHEEVADLTSGASGEVEEKLDALEEVVISIVTESDLFSDMN 679
S A + VDE+ D EE D T E EEK
Sbjct: 657 EKSEADNDREDEVVDEEKDKEEEGDDETENEDKEDEEK---------------------- 770
Query: 680 SGASEEVEEKEGRRTADKGHGVDVATSYLTGSVDADLSELVPASTLLDSEQAVNTEANNL 739
G E E E R K A ++ T + L + L + + E N+
Sbjct: 771 GGLVENHENHEAREEHYKADDASSAVAHDTHETSTETGNLEHSDLSLQNTRKPENETNHS 950
Query: 740 TTSLESELVANDEETTHDIVD 760
S S+ V++ + T ++ D
Sbjct: 951 DESYGSQNVSDLKVTEGELTD 1013
>TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protein {Oryza
sativa (japonica cultivar-group)}, partial (62%)
Length = 2345
Score = 32.7 bits (73), Expect = 0.80
Identities = 42/171 (24%), Positives = 66/171 (38%), Gaps = 6/171 (3%)
Frame = +1
Query: 305 EVEEEVDGLAVSAATQELDMFGDLNSGGALGEV----EEKLDGDEDDAISTVTPESDMFS 360
EV E+V S A Q D GDL G+ EEK + + + S T D
Sbjct: 487 EVREQVSETDNSNARQFEDNPGDLPEDATKGDSNVSSEEKSEENSTEKSSEDTKTEDEGK 666
Query: 361 DLNNGASGEVEEKLDGDEEDAISTVTPESDMF--SDMNNGASGEVEEKLDFHEADALSTD 418
+ S E DG+E + + ES+ S+ NN + + EK + S +
Sbjct: 667 KTEDEGSN-TENNKDGEEASTKESESDESEKKDESEENNKSDSDESEK-----KSSDSNE 828
Query: 419 TPGSNVNSDMNNGASGEVEEKIDDHEEASVSSITPESDMFTDISSGALGEV 469
T SNV + + E +E + + +++F SGA E+
Sbjct: 829 TTDSNVEEKVEQSQNKESDENASEKNTDDNAKDQSSNEVF---PSGAQSEL 972
>TC80941 weakly similar to PIR|T39903|T39903 serine-rich protein - fission
yeast (Schizosaccharomyces pombe), partial (15%)
Length = 557
Score = 32.7 bits (73), Expect = 0.80
Identities = 18/58 (31%), Positives = 29/58 (49%)
Frame = +2
Query: 333 ALGEVEEKLDGDEDDAISTVTPESDMFSDLNNGASGEVEEKLDGDEEDAISTVTPESD 390
A G +EEK +G++++ E D N + E EEK +GDE++ T E +
Sbjct: 143 AEGSIEEKEEGNDEEKTEVEEKEE---GDDNEKSEEETEEKEEGDEKEKTEAETEEEE 307
>TC93337 weakly similar to GP|21240660|gb|AAM44371.1 hypothetical protein
{Dictyostelium discoideum}, partial (7%)
Length = 743
Score = 32.3 bits (72), Expect = 1.0
Identities = 36/189 (19%), Positives = 76/189 (40%), Gaps = 6/189 (3%)
Frame = +1
Query: 649 GASGEVEEKLDALEEVVISIVTESDLFSDMNSGASEEVEEKEGRRTADKGHGVDVATSYL 708
G G ++++D + +V S+ F D EE +EKE + D+G+ V ++ +
Sbjct: 64 GERGGEDDEMDENDPEKPEVVDRSEEFVDEGKEKEEEGDEKENENSKDEGNQGLVESNNI 243
Query: 709 TGSVDADLSELVPASTLLDSEQAVNTEANNLT---TSLESELVANDEETTHDIVDDSI-- 763
+ + +S + + +TE ++ + + +E+ E + +D I
Sbjct: 244 HEAREEQYKGDDASSAVTHDTRTTSTETETVSMENSDVNAEMGITKPENKPNYTEDGIRS 423
Query: 764 -DASEMEKSTMLHDLPASSDMENKIDVGNTERNDYESSSHLTVPELHSVEMASYGENTSR 822
S + + + A S+ + + GN ++ + SH L + + E TS
Sbjct: 424 QPGSNFSITEVKLVVGAYSNSSSSNETGNNSLSNPVAGSHQNNTAL--IYSVRHSEETSS 597
Query: 823 TELFLVSAA 831
+V AA
Sbjct: 598 NLTIVVPAA 624
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.307 0.127 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,323,623
Number of Sequences: 36976
Number of extensions: 340532
Number of successful extensions: 1736
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 1607
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1710
length of query: 1113
length of database: 9,014,727
effective HSP length: 106
effective length of query: 1007
effective length of database: 5,095,271
effective search space: 5130937897
effective search space used: 5130937897
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0314.17


