

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
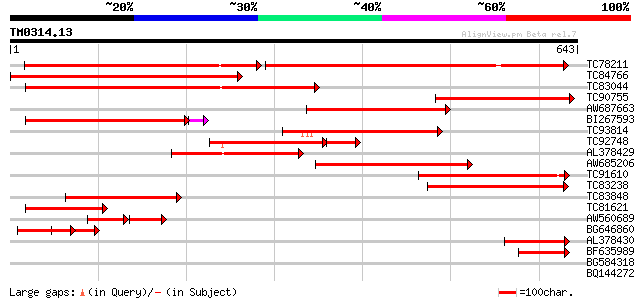
Query= TM0314.13
(643 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78211 weakly similar to GP|15624010|dbj|BAB68064. sulfate tran... 293 e-145
TC84766 similar to SP|P53392|SUT2_STYHA High affinity sulphate t... 400 e-112
TC83044 similar to PIR|T48901|T48901 sulfate transporter ATST1 [... 364 e-101
TC90755 similar to SP|P53391|SUT1_STYHA High affinity sulphate t... 269 3e-72
AW687663 homologue to GP|13487717|gb sulfate transporter 2 {Lyco... 240 1e-63
BI267593 similar to PIR|B86365|B86 probable sulphate transporter... 208 3e-54
TC93814 similar to PIR|B86365|B86365 probable sulphate transport... 192 3e-49
TC92748 similar to SP|P53393|SUT3_STYHA Low affinity sulphate tr... 127 1e-38
AL378429 similar to GP|9294446|dbj sulfate transporter {Arabidop... 154 8e-38
AW685206 similar to GP|9955547|emb sulfate transporter {Arabidop... 150 1e-36
TC91610 weakly similar to PIR|T50022|T50022 sulfate transporter ... 148 7e-36
TC83238 weakly similar to PIR|T49069|T49069 sulfate transporter ... 141 7e-34
TC83848 similar to SP|P53393|SUT3_STYHA Low affinity sulphate tr... 134 1e-31
TC81621 similar to PIR|T49069|T49069 sulfate transporter (ATST1)... 105 5e-23
AW560689 similar to GP|9294446|dbj| sulfate transporter {Arabido... 59 7e-17
BG646860 similar to GP|13487717|gb| sulfate transporter 2 {Lycop... 76 5e-14
AL378430 similar to GP|9294446|dbj| sulfate transporter {Arabido... 67 2e-11
BF635989 weakly similar to GP|9294446|dbj| sulfate transporter {... 48 1e-05
BG584318 similar to PIR|T39116|T391 probable sulfate permease - ... 38 0.011
BQ144272 similar to GP|13487715|gb| sulfate transporter 1 {Lycop... 35 0.12
>TC78211 weakly similar to GP|15624010|dbj|BAB68064. sulfate transporter-like
protein {Oryza sativa (japonica cultivar-group)}, partial
(66%)
Length = 2090
Score = 293 bits (749), Expect(2) = e-145
Identities = 144/343 (41%), Positives = 233/343 (66%)
Frame = +2
Query: 291 YITRADKKGVAIVKHIKKGVNPASASEIFFSGEYFGAGVKIGVVSGMVALTEAVAIARTF 350
Y+ ++ + G+ IV H+ KG+NP S + F Y +K G++SG+++L E +AI R+F
Sbjct: 893 YLVKSTQHGIQIVGHLDKGLNPISIQFLTFDRRYLSTVMKAGLISGVLSLAEGIAIGRSF 1072
Query: 351 AAMKDYSIDGNKEMVAMGTMNIVGSLTSSYVATGSFSRSAVNYMAGCKTAVSNIVMSMVL 410
+ + DGNKEM+A G MN+VGS TS Y+ +G FS++AVNY AGCK+A++N+V ++++
Sbjct: 1073 SVTANTPHDGNKEMIAFGLMNLVGSFTSCYLTSGPFSKTAVNYNAGCKSAMTNVVQAVIM 1252
Query: 411 LLTLLVITPLFKYTPNAVLASIIIAAVMNLVDVQAAILLWKIDKFDFVACMGAFFGVIFK 470
LTL ++ PLF TP L++II++A++ L++ AI L+K+DKFDF+ CM AF GV F
Sbjct: 1253 ALTLQLLAPLFSNTPLVALSAIIVSAMLGLINYTEAIHLFKVDKFDFIICMSAFLGVAFL 1432
Query: 471 SVEIGLLIAVAISFAKILLQVTRPRTAILGKLSGTKVYRNILQYPKATQIPGMLIIRVDS 530
S++IGL+++V + + LL + RP LGKL + +YR++ QY A+ IPG+LII+V S
Sbjct: 1433 SMDIGLMLSVGLGVLRGLLYLARPPACKLGKLPDSGLYRDVEQYSNASTIPGVLIIQVGS 1612
Query: 531 AIYFSNSNYIKDRILKWVTDEEVQRTASEFPSIQSLIVEMSPVTDIDTSGIHALEDLFKS 590
IYFSNS Y+K+RIL+++ E+ +S ++ +I+ ++ V+ IDT+ I L + K
Sbjct: 1613 PIYFSNSTYLKERILRYIKSEQ----SSSGDMVEHVILVLTAVSSIDTTAIEGLLETQKI 1780
Query: 591 LKKREVQLLLANPGPIVIEKLHASKLSDIIGEDKIFSSVDDAV 633
L+ + +Q+ L NP V+EKL ASK + +G++ + +++DAV
Sbjct: 1781 LEMKGIQMALVNPRLEVMEKLIASKFVEKVGKESFYLNLEDAV 1909
Score = 242 bits (618), Expect(2) = e-145
Identities = 127/270 (47%), Positives = 174/270 (64%), Gaps = 2/270 (0%)
Frame = +1
Query: 18 HIHKVGTAPKQTLYLEIKHSVKETFFFDDPLSQFKGQSRK-RKLVLGMQSVFPIVEWGRD 76
H + V + ++ ++K KE F DDP Q + +K R+L+ G+Q PI EW +
Sbjct: 70 HHNGVNLSTQRGFVTKLKSGFKEALFPDDPFRQIMEEEKKSRRLIKGVQYFIPIFEWLPN 249
Query: 77 YNLQKFKGDFIAGLTIASLCIPQDIAYAKLANLNPEHALYTSFVAPLVYAFMGTSRDIAI 136
Y+L+ F D IAGLTIASL IPQ I+YAKLANL P LY+SFV PLVYA G+SR +A+
Sbjct: 250 YSLRLFFSDLIAGLTIASLAIPQGISYAKLANLPPLIGLYSSFVPPLVYAVFGSSRHMAV 429
Query: 137 GPVAVVSLLLGTMLTDEIADSKNPD-YLRLAYTATFFAGITQLALGFFRLGFLIDFLSHA 195
G +A SLL+ ++++ P YL L +T TF G+ Q LGFFRLG L+DF SH+
Sbjct: 430 GTIAAASLLIASIVSTVADPIAEPTLYLHLIFTTTFITGVFQACLGFFRLGILVDFFSHS 609
Query: 196 AIVGFMGGAAITIALQQLKGLLGLKTFTKKTDIVSVMRSVWKPVEHGWNWETIVIGMSFL 255
I GFMGG A+ + LQQ KG+ G+K F+ KT++V+V+ ++ H WET V+G+ FL
Sbjct: 610 TITGFMGGTAVILILQQFKGIFGMKHFSTKTNVVAVLEGIFSN-RHEIRWETTVLGIIFL 786
Query: 256 VFILITNYIAKKNKKLFWVAAIAPMISVVV 285
VF+ T ++ K KLFWV+AIAP+ VVV
Sbjct: 787 VFLQFTRHLRLKKPKLFWVSAIAPITCVVV 876
>TC84766 similar to SP|P53392|SUT2_STYHA High affinity sulphate transporter
2. {Stylosanthes hamata}, partial (37%)
Length = 873
Score = 400 bits (1028), Expect = e-112
Identities = 197/266 (74%), Positives = 230/266 (86%), Gaps = 2/266 (0%)
Frame = +1
Query: 1 MEDTGSAPSSRRHHG--LPHIHKVGTAPKQTLYLEIKHSVKETFFFDDPLSQFKGQSRKR 58
ME+ S PS R H G LPH+HKV PKQTL+ + KHS ETFF DDP ++FK Q++KR
Sbjct: 37 MEEIKSNPS*RGHGGAILPHMHKVSGPPKQTLFQDFKHSFNETFFSDDPFAKFKDQTKKR 216
Query: 59 KLVLGMQSVFPIVEWGRDYNLQKFKGDFIAGLTIASLCIPQDIAYAKLANLNPEHALYTS 118
K VLG+QSVFPI+EWGR YNL+ FKGD I+GLTIASLCIPQDIAYAKLANL P++ALYTS
Sbjct: 217 KFVLGLQSVFPILEWGRGYNLKSFKGDLISGLTIASLCIPQDIAYAKLANLEPQYALYTS 396
Query: 119 FVAPLVYAFMGTSRDIAIGPVAVVSLLLGTMLTDEIADSKNPDYLRLAYTATFFAGITQL 178
FVAPLVYAFMG+SRDIAIGPVAVVSLLLG++L++EI+D K+P+YL LA+T+TFFAG+ Q+
Sbjct: 397 FVAPLVYAFMGSSRDIAIGPVAVVSLLLGSLLSEEISDFKSPEYLALAFTSTFFAGVVQM 576
Query: 179 ALGFFRLGFLIDFLSHAAIVGFMGGAAITIALQQLKGLLGLKTFTKKTDIVSVMRSVWKP 238
ALG +LGFLIDFLSHAAIVGFMGGAAITIALQQLKGLLG+K FTKKTDIVSVM SV+K
Sbjct: 577 ALGVLKLGFLIDFLSHAAIVGFMGGAAITIALQQLKGLLGIKKFTKKTDIVSVMTSVFKA 756
Query: 239 VEHGWNWETIVIGMSFLVFILITNYI 264
HGWNW+TI+IG+SFL F+ IT YI
Sbjct: 757 AHHGWNWQTIIIGLSFLGFLFITKYI 834
>TC83044 similar to PIR|T48901|T48901 sulfate transporter ATST1 [imported] -
Arabidopsis thaliana, partial (50%)
Length = 1278
Score = 364 bits (934), Expect = e-101
Identities = 177/334 (52%), Positives = 236/334 (69%), Gaps = 1/334 (0%)
Frame = +1
Query: 19 IHKVGTAPKQTLYLEIKHSVKETFFFDDPLSQFKGQSRKRKLVLGMQSVFPIVEWGRDYN 78
+H V P Q +K+S+KETFF DDPL +FK Q +KLVLG+Q FPI EW Y
Sbjct: 274 VHGVAIPPPQPFLKSMKYSMKETFFPDDPLRRFKNQPASKKLVLGLQYFFPIFEWAPSYT 453
Query: 79 LQKFKGDFIAGLTIASLCIPQDIAYAKLANLNPEHALYTSFVAPLVYAFMGTSRDIAIGP 138
K D IAG+TIASL IPQ I+YAKLANL P LY+SF+ PL+YA MG+SRD+A+G
Sbjct: 454 FHFLKSDLIAGITIASLAIPQGISYAKLANLPPILGLYSSFIPPLIYAMMGSSRDLAVGT 633
Query: 139 VAVVSLLLGTMLTDEIADSKNPD-YLRLAYTATFFAGITQLALGFFRLGFLIDFLSHAAI 197
VAV SLL+G+ML +E+ ++NP +L LA+TATFFAG+ Q +LG FRLGF++DFLSHAAI
Sbjct: 634 VAVGSLLMGSMLANEVNPTQNPKLFLHLAFTATFFAGLLQASLGLFRLGFIVDFLSHAAI 813
Query: 198 VGFMGGAAITIALQQLKGLLGLKTFTKKTDIVSVMRSVWKPVEHGWNWETIVIGMSFLVF 257
VGFMGGAA + LQQLK +LGL+ FT DIVSVMRSV+ H W WE+ V+G F+ F
Sbjct: 814 VGFMGGAATVVCLQQLKSILGLEHFTHAADIVSVMRSVFTQT-HQWRWESAVLGFCFIFF 990
Query: 258 ILITNYIAKKNKKLFWVAAIAPMISVVVSTFCVYITRADKKGVAIVKHIKKGVNPASASE 317
+L+T Y +KK K FWV+A+ P+ SV++ + VY T A+ GV ++ +KKG+NP S ++
Sbjct: 991 LLVTRYFSKKQPKFFWVSAMTPLASVILGSLLVYFTHAEHHGVQVIGELKKGLNPPSLTD 1170
Query: 318 IFFSGEYFGAGVKIGVVSGMVALTEAVAIARTFA 351
+ F Y +K G++ G++AL E +A+ R+FA
Sbjct: 1171LVFVSPYMTTAIKTGLIVGIIALAEGIAVGRSFA 1272
>TC90755 similar to SP|P53391|SUT1_STYHA High affinity sulphate transporter
1. {Stylosanthes hamata}, partial (24%)
Length = 847
Score = 269 bits (687), Expect = 3e-72
Identities = 133/157 (84%), Positives = 146/157 (92%)
Frame = +1
Query: 484 FAKILLQVTRPRTAILGKLSGTKVYRNILQYPKATQIPGMLIIRVDSAIYFSNSNYIKDR 543
F + LLQVTRP+TA+LGKL GT VYRNILQYPKA QIPGMLI+RVDSAIYFSNSNYIKDR
Sbjct: 22 FCQNLLQVTRPKTAVLGKLPGTTVYRNILQYPKAAQIPGMLIVRVDSAIYFSNSNYIKDR 201
Query: 544 ILKWVTDEEVQRTASEFPSIQSLIVEMSPVTDIDTSGIHALEDLFKSLKKREVQLLLANP 603
ILKW+TDEE+ RT+SE+PSIQ LIVEMSPVTDIDTSGIH+ EDL KSLKKR++QLLLANP
Sbjct: 202 ILKWLTDEEILRTSSEYPSIQHLIVEMSPVTDIDTSGIHSFEDLLKSLKKRDIQLLLANP 381
Query: 604 GPIVIEKLHASKLSDIIGEDKIFSSVDDAVATFGPKG 640
GPIVIEKLHASKLSD+IGEDKIF +V DAVATFGPKG
Sbjct: 382 GPIVIEKLHASKLSDLIGEDKIFLTVGDAVATFGPKG 492
>AW687663 homologue to GP|13487717|gb sulfate transporter 2 {Lycopersicon
esculentum}, partial (23%)
Length = 526
Score = 240 bits (613), Expect = 1e-63
Identities = 123/164 (75%), Positives = 142/164 (86%), Gaps = 1/164 (0%)
Frame = +1
Query: 337 MVALTEAVAIARTFAAMKDYSIDGNKEMVAMGTMNIVGSLTSSYVATGSFSRSAVNYMAG 396
M+ALTEA+AI RTFA+MKDY +DGNKEMVA+G MN+VGS+TS YVATGSFSRSAVN+MAG
Sbjct: 1 MIALTEAIAIGRTFASMKDYQLDGNKEMVALGAMNVVGSMTSCYVATGSFSRSAVNFMAG 180
Query: 397 CKTAVSNIVMSMVLLLTLLVITPLFKYTPNAVLASIIIAAVMNLVDVQAAILLWKIDKFD 456
C+TAVSNIVMS+V+ LTL ITPLFKYTPNA+LASIII AV+NLVD +AAIL+WKIDKFD
Sbjct: 181 CETAVSNIVMSVVVFLTLQFITPLFKYTPNAILASIIICAVINLVDYKAAILIWKIDKFD 360
Query: 457 FVACMGAFFGVIFKSVEIGLLIAVAI-SFAKILLQVTRPRTAIL 499
FVACMGAFFGV+F SVEIGLLIA + + + PRTAIL
Sbjct: 361 FVACMGAFFGVVFASVEIGLLIACFLYPLQRSYCKSQGPRTAIL 492
Score = 29.3 bits (64), Expect = 4.9
Identities = 18/30 (60%), Positives = 20/30 (66%), Gaps = 1/30 (3%)
Frame = +3
Query: 481 AISFAKILLQVTR-PRTAILGKLSGTKVYR 509
+ISFAKILLQVTR P G+ T VYR
Sbjct: 435 SISFAKILLQVTRAPNGYSRGRSLRTTVYR 524
>BI267593 similar to PIR|B86365|B86 probable sulphate transporter protein
[imported] - Arabidopsis thaliana, partial (32%)
Length = 646
Score = 208 bits (529), Expect(2) = 3e-54
Identities = 102/187 (54%), Positives = 141/187 (74%), Gaps = 1/187 (0%)
Frame = +3
Query: 19 IHKVGTAPKQTLYLEIKHSVKETFFFDDPLSQFKGQSRKRKLVLGMQSVFPIVEWGRDYN 78
IH+V P ++ + K +KETFF DDPL QFKGQ+ +++L+LG + FPI++WG Y+
Sbjct: 21 IHQVVPPPHKSTLQKFKVRLKETFFPDDPLRQFKGQTLQKRLILGAKYFFPILQWGSIYS 200
Query: 79 LQKFKGDFIAGLTIASLCIPQDIAYAKLANLNPEHALYTSFVAPLVYAFMGTSRDIAIGP 138
+ FK D ++GLTIASL IPQ I+YAKLANL P LY+SFV PL+YA +G+SRD+A+GP
Sbjct: 201 FKLFKSDLVSGLTIASLAIPQGISYAKLANLPPIVGLYSSFVPPLIYAVLGSSRDLAVGP 380
Query: 139 VAVVSLLLGTMLTDEIADSKNPD-YLRLAYTATFFAGITQLALGFFRLGFLIDFLSHAAI 197
V++ SL+LG+ML E++ S P +L+LA T+TFFAG+ Q +LG RLGF+IDFLS A +
Sbjct: 381 VSIASLVLGSMLRQEVSPSAEPVLFLQLALTSTFFAGLFQASLGILRLGFIIDFLSKAIL 560
Query: 198 VGFMGGA 204
+GFM G+
Sbjct: 561 IGFMAGS 581
Score = 22.7 bits (47), Expect(2) = 3e-54
Identities = 12/23 (52%), Positives = 13/23 (56%)
Frame = +1
Query: 203 GAAITIALQQLKGLLGLKTFTKK 225
GAAI QLK LLG+ T K
Sbjct: 577 GAAIIGVFAQLKSLLGITHXTHK 645
>TC93814 similar to PIR|B86365|B86365 probable sulphate transporter protein
[imported] - Arabidopsis thaliana, partial (30%)
Length = 671
Score = 192 bits (489), Expect = 3e-49
Identities = 101/202 (50%), Positives = 140/202 (69%), Gaps = 20/202 (9%)
Frame = +2
Query: 310 VNPASASEIFFSGEYFGAGVK-----------IGVVSG----MVAL-----TEAVAIART 349
+NP+S + + F G + G VK +G ++G ++ L E +A+ RT
Sbjct: 2 INPSSWNMLKFHGSHLGLVVKTWHYHWHFVPHLGFINGKNG*VICLFKKNKQEGIAVGRT 181
Query: 350 FAAMKDYSIDGNKEMVAMGTMNIVGSLTSSYVATGSFSRSAVNYMAGCKTAVSNIVMSMV 409
FAA+ +Y +DGNKEM+A+G MN+VGS TS YV TG+FSRSAVN AG KTA SNIVMS+
Sbjct: 182 FAALGNYKVDGNKEMMAIGCMNVVGSFTSCYVTTGAFSRSAVNNNAGAKTAASNIVMSVT 361
Query: 410 LLLTLLVITPLFKYTPNAVLASIIIAAVMNLVDVQAAILLWKIDKFDFVACMGAFFGVIF 469
+++TLL + PLF+YTPN VL +II+ AV+ L+D+ AA +WKIDKFDF+ + AFFGV+F
Sbjct: 362 VMVTLLFLMPLFQYTPNVVLGAIIVTAVIGLIDIPAACHIWKIDKFDFLVMLTAFFGVVF 541
Query: 470 KSVEIGLLIAVAISFAKILLQV 491
SV++GL +AV +S ILLQ+
Sbjct: 542 ISVQLGLAVAVGLSTFXILLQI 607
>TC92748 similar to SP|P53393|SUT3_STYHA Low affinity sulphate transporter
3. {Stylosanthes hamata}, partial (20%)
Length = 536
Score = 127 bits (320), Expect(2) = 1e-38
Identities = 64/141 (45%), Positives = 99/141 (69%), Gaps = 7/141 (4%)
Frame = +2
Query: 227 DIVSVMRSVWKPV------EHGWNWETIVIGMSFLVFILITNYIAKKNKKLFWVAAIAPM 280
D VSV+ SV+K + E W+ V+G SFL+F+L+T +IA+K KKLFW+ AIAP+
Sbjct: 2 DAVSVLVSVYKSLHQQITSEEKWSPLNFVLGCSFLIFLLVTRFIARKKKKLFWLPAIAPL 181
Query: 281 ISVVVSTFCVYITRADKKGVAIVKHIKKGVNPASASEIFFSGEYFGAGVKIGVVSGMVAL 340
+SV++ST VY+++ADK+G+ I+KH+K G+N +S ++ F G+ G KIG+V ++AL
Sbjct: 182 LSVILSTLIVYLSKADKQGINIIKHVKGGLNQSSVHQLQFHGQNVGQAAKIGLVCAVIAL 361
Query: 341 TEAVAIARTFAAMK-DYSIDG 360
TEA+A +FA ++ D +DG
Sbjct: 362 TEAMAGGPSFAFLQXDTHLDG 424
Score = 51.2 bits (121), Expect(2) = 1e-38
Identities = 25/38 (65%), Positives = 28/38 (72%)
Frame = +3
Query: 360 GNKEMVAMGTMNIVGSLTSSYVATGSFSRSAVNYMAGC 397
GN+EMVAMG MN G+LTS YV GS SR+AV M GC
Sbjct: 423 GNREMVAMGIMNXAGTLTSWYVXPGSXSRTAVISMXGC 536
>AL378429 similar to GP|9294446|dbj sulfate transporter {Arabidopsis
thaliana}, partial (22%)
Length = 473
Score = 154 bits (390), Expect = 8e-38
Identities = 72/150 (48%), Positives = 105/150 (70%)
Frame = +3
Query: 184 RLGFLIDFLSHAAIVGFMGGAAITIALQQLKGLLGLKTFTKKTDIVSVMRSVWKPVEHGW 243
RLGF+IDFLS A +VGFM GAAI ++LQQLKGLLG+ FT K I+ V+ SV+K + W
Sbjct: 27 RLGFVIDFLSKATLVGFMAGAAIIVSLQQLKGLLGIVHFTNKMQIIPVLVSVYKQKDE-W 203
Query: 244 NWETIVIGMSFLVFILITNYIAKKNKKLFWVAAIAPMISVVVSTFCVYITRADKKGVAIV 303
+W+TIV+G FL F+L T +I+ + KLFW +A AP+ SV++ST V++ R ++++
Sbjct: 204 SWQTIVMGFGFLAFLLTTRHISLRKPKLFWASAAAPLTSVILSTILVFLLRNKAHHISVI 383
Query: 304 KHIKKGVNPASASEIFFSGEYFGAGVKIGV 333
H+ KGVNP S + ++F+G Y +K G+
Sbjct: 384 GHLAKGVNPPSVNLLYFNGPYLALAIKTGI 473
>AW685206 similar to GP|9955547|emb sulfate transporter {Arabidopsis
thaliana}, partial (25%)
Length = 540
Score = 150 bits (380), Expect = 1e-36
Identities = 75/178 (42%), Positives = 113/178 (63%)
Frame = +3
Query: 348 RTFAAMKDYSIDGNKEMVAMGTMNIVGSLTSSYVATGSFSRSAVNYMAGCKTAVSNIVMS 407
+ AA Y +D N+E+V +G N++GS S+Y TGSFSRSAVN+ +G K+ VS IV
Sbjct: 3 KALAAKNGYELDSNQELVGLGVSNVLGSFFSAYPTTGSFSRSAVNHESGAKSGVSAIVSG 182
Query: 408 MVLLLTLLVITPLFKYTPNAVLASIIIAAVMNLVDVQAAILLWKIDKFDFVACMGAFFGV 467
+++ LL +TPLF+ P + LA+I+I+AV+ LVD AI LW++DK DF+ +
Sbjct: 183 IIITCALLFLTPLFENIPQSALAAIVISAVIGLVDYDEAIFLWRVDKKDFLLWILTSTTT 362
Query: 468 IFKSVEIGLLIAVAISFAKILLQVTRPRTAILGKLSGTKVYRNILQYPKATQIPGMLI 525
+F +EIG+++ V S A ++ + P A+LG+L GT V RN QY +A G++I
Sbjct: 363 LFLGIEIGVMVGVGASLAFVIHESANPHIAVLGRLPGTTVXRNXKQYXEAYTYNGIVI 536
>TC91610 weakly similar to PIR|T50022|T50022 sulfate transporter -
Arabidopsis thaliana, partial (21%)
Length = 756
Score = 148 bits (373), Expect = 7e-36
Identities = 83/174 (47%), Positives = 121/174 (68%), Gaps = 2/174 (1%)
Frame = +3
Query: 464 FFGVIFKSVEIGLLIAVAISFAKILLQVTRPRTAILGKLSGTKVYRNILQYPKATQIPGM 523
FFGV+F SVEIGLL+AV ISFAKI++ RP T LGKL GT ++ ++ QYP A QIPG+
Sbjct: 3 FFGVLFASVEIGLLVAVVISFAKIIVISIRPSTETLGKLPGTDLFCDVDQYPMAIQIPGV 182
Query: 524 LIIRVDSAIY-FSNSNYIKDRILKWVTDEEVQRTASEFPS-IQSLIVEMSPVTDIDTSGI 581
+IIR+ SA+ F+N+N++K+RI+KWVT + ++ S IQ +I++ S + +IDTSGI
Sbjct: 183 MIIRMKSALLCFANANFVKERIIKWVTQKGLEDDKGNSKSTIQLVILDTSNLVNIDTSGI 362
Query: 582 HALEDLFKSLKKREVQLLLANPGPIVIEKLHASKLSDIIGEDKIFSSVDDAVAT 635
++E+L+K L QL +ANP VI KL S IG +++ +V++AVA+
Sbjct: 363 ASMEELYKCLSTHGKQLAIANPRWQVIHKLKVSNFVSKIG-GRVYLTVEEAVAS 521
>TC83238 weakly similar to PIR|T49069|T49069 sulfate transporter (ATST1) -
Arabidopsis thaliana, partial (21%)
Length = 778
Score = 141 bits (356), Expect = 7e-34
Identities = 67/160 (41%), Positives = 112/160 (69%)
Frame = +2
Query: 474 IGLLIAVAISFAKILLQVTRPRTAILGKLSGTKVYRNILQYPKATQIPGMLIIRVDSAIY 533
IGL+IAVAIS ++LL V RPRT +LG + + YRN+ Y A +PG+LI+ +D+ I+
Sbjct: 2 IGLIIAVAISVLRVLLFVARPRTFVLGNIPNSGAYRNVEHYQNAHHVPGILILEIDAPIF 181
Query: 534 FSNSNYIKDRILKWVTDEEVQRTASEFPSIQSLIVEMSPVTDIDTSGIHALEDLFKSLKK 593
F+N++Y+++RI +W+ +EE + + S+Q +I++M+ V +IDTSGI LE+ K++ +
Sbjct: 182 FANASYLRERITRWINEEEERIEGAGKTSLQYVIMDMTAVANIDTSGISMLEEFKKTVDR 361
Query: 594 REVQLLLANPGPIVIEKLHASKLSDIIGEDKIFSSVDDAV 633
+ +QL++ NPG V +KL+ SK D IG ++ +V++AV
Sbjct: 362 KGLQLVMVNPGSEVTKKLNKSKFLDEIGHKWVYVTVEEAV 481
>TC83848 similar to SP|P53393|SUT3_STYHA Low affinity sulphate transporter
3. {Stylosanthes hamata}, partial (21%)
Length = 769
Score = 134 bits (336), Expect = 1e-31
Identities = 70/134 (52%), Positives = 93/134 (69%), Gaps = 2/134 (1%)
Frame = +3
Query: 64 MQSVFPIVEWGRDYNLQKFKGDFIAGLTIASLCIPQDIAYAKLANLNPEHALYTSFVAPL 123
+QS+FPI+ W +DY + KFK D +AGLT+ASLCIPQ I YA LA ++P++ LYTS V PL
Sbjct: 333 LQSLFPILVWLKDYTISKFKDDLLAGLTLASLCIPQSIGYASLAKVDPQYGLYTSIVPPL 512
Query: 124 VYAFMGTSRDIAIGPVAVVSLLLGTMLTDEIADSKNPD-YLRLAYTATFFAGITQLALGF 182
+YA MG+SRDIAIGPVAVVS+LL +++T+ I NP Y +T + +
Sbjct: 513 IYAVMGSSRDIAIGPVAVVSMLLSSLVTNVIDPVANPHAYRDFIFTVHLLHRNFSSCIWY 692
Query: 183 FRLGF-LIDFLSHA 195
F++GF L FLSHA
Sbjct: 693 FQVGFSLWIFLSHA 734
Score = 37.4 bits (85), Expect = 0.018
Identities = 18/39 (46%), Positives = 23/39 (58%)
Frame = +1
Query: 165 LAYTATFFAGITQLALGFFRLGFLIDFLSHAAIVGFMGG 203
L+ +TFF GI Q A G FRLGFL F H + ++ G
Sbjct: 640 LSLPSTFFTGIFQAAFGIFRLGFLCGFFFHMPALVWIHG 756
>TC81621 similar to PIR|T49069|T49069 sulfate transporter (ATST1) -
Arabidopsis thaliana, partial (14%)
Length = 863
Score = 105 bits (262), Expect = 5e-23
Identities = 51/93 (54%), Positives = 62/93 (65%)
Frame = +2
Query: 19 IHKVGTAPKQTLYLEIKHSVKETFFFDDPLSQFKGQSRKRKLVLGMQSVFPIVEWGRDYN 78
+H+V P Q + +K+SVKET F DDPL +FK Q R+ VL +Q PI+EW Y
Sbjct: 461 VHRVAIPPHQPFFESLKYSVKETLFPDDPLRKFKNQPASRRFVLALQHFLPILEWAPQYT 640
Query: 79 LQKFKGDFIAGLTIASLCIPQDIAYAKLANLNP 111
LQ K D IAG+TIASL IPQ I+YAKLANL P
Sbjct: 641 LQFLKSDLIAGITIASLAIPQGISYAKLANLPP 739
>AW560689 similar to GP|9294446|dbj| sulfate transporter {Arabidopsis
thaliana}, partial (13%)
Length = 276
Score = 58.5 bits (140), Expect(2) = 7e-17
Identities = 29/46 (63%), Positives = 35/46 (76%)
Frame = +3
Query: 89 GLTIASLCIPQDIAYAKLANLNPEHALYTSFVAPLVYAFMGTSRDI 134
GLTIASL IPQ I+YAKLANL P LY+S V P +YA +G+SR +
Sbjct: 3 GLTIASLAIPQGISYAKLANLPPIIGLYSSIVPP*IYALLGSSRHV 140
Score = 47.0 bits (110), Expect(2) = 7e-17
Identities = 21/43 (48%), Positives = 35/43 (80%), Gaps = 1/43 (2%)
Frame = +1
Query: 136 IGPVAVVSLLLGTMLTDEIADSKNPD-YLRLAYTATFFAGITQ 177
+GPV++ SL++G+ML++ + ++NP YL+LA+TATF AG+ Q
Sbjct: 145 VGPVSIASLVMGSMLSETVFFTQNPTLYLQLAFTATFVAGLFQ 273
>BG646860 similar to GP|13487717|gb| sulfate transporter 2 {Lycopersicon
esculentum}, partial (13%)
Length = 752
Score = 75.9 bits (185), Expect = 5e-14
Identities = 33/65 (50%), Positives = 45/65 (68%)
Frame = +2
Query: 10 SRRHHGLPHIHKVGTAPKQTLYLEIKHSVKETFFFDDPLSQFKGQSRKRKLVLGMQSVFP 69
S H P+ +KV PKQ L+ E +++VKETFF DDPL FK QS +KL+LG++ +FP
Sbjct: 302 SASHDQQPYAYKVAIPPKQNLFKEFQYTVKETFFADDPLRSFKDQSTSKKLILGIEFIFP 481
Query: 70 IVEWG 74
I+ WG
Sbjct: 482 ILNWG 496
Score = 53.5 bits (127), Expect = 2e-07
Identities = 30/54 (55%), Positives = 37/54 (67%)
Frame = +3
Query: 48 LSQFKGQSRKRKLVLGMQSVFPIVEWGRDYNLQKFKGDFIAGLTIASLCIPQDI 101
LS+ K + LVL S++ + GR YNL+KF+GD IAGLTIASLCIPQ I
Sbjct: 423 LSKTKVHQKS*YLVLSSYSLY--LTGGRSYNLKKFRGDIIAGLTIASLCIPQVI 578
>AL378430 similar to GP|9294446|dbj| sulfate transporter {Arabidopsis
thaliana}, partial (13%)
Length = 475
Score = 67.4 bits (163), Expect = 2e-11
Identities = 35/73 (47%), Positives = 51/73 (68%)
Frame = +2
Query: 562 SIQSLIVEMSPVTDIDTSGIHALEDLFKSLKKREVQLLLANPGPIVIEKLHASKLSDIIG 621
S+Q +I++M+ VT IDTSG+ L +L K L+KR +QL+L NP V+EKLH SK+ D G
Sbjct: 23 SLQCIILDMTAVTAIDTSGLETLGELRKMLEKRSLQLVLVNPVGNVMEKLHMSKILDTFG 202
Query: 622 EDKIFSSVDDAVA 634
++ +V +AVA
Sbjct: 203 FKGLYLTVGEAVA 241
>BF635989 weakly similar to GP|9294446|dbj| sulfate transporter {Arabidopsis
thaliana}, partial (9%)
Length = 655
Score = 48.1 bits (113), Expect = 1e-05
Identities = 26/57 (45%), Positives = 38/57 (66%)
Frame = +2
Query: 578 TSGIHALEDLFKSLKKREVQLLLANPGPIVIEKLHASKLSDIIGEDKIFSSVDDAVA 634
TSGI L +L + L+++ +QL+LANP V+EKLH S + D G ++ SV +AVA
Sbjct: 2 TSGIDTLCELRRRLEQKSLQLVLANPVGNVMEKLHESNILDSFGMKGLYLSVGEAVA 172
>BG584318 similar to PIR|T39116|T391 probable sulfate permease - fission
yeast (Schizosaccharomyces pombe), partial (4%)
Length = 811
Score = 38.1 bits (87), Expect = 0.011
Identities = 29/131 (22%), Positives = 62/131 (47%), Gaps = 27/131 (20%)
Frame = -3
Query: 521 PGMLIIRVDSAIYFSNSNYIKDRILKWVTD--EEVQRTAS-------------------- 558
PG++I ++ ++ + N+N++ D+I+ + + ++Q A
Sbjct: 578 PGVIIFQLGESLIYVNANHVLDKIINYTKENTRDMQHKAEKKGDRPWNDSKNESAAKQSQ 399
Query: 559 --EFPSIQSLIVEMSPVTDIDTSGIHALEDLFKSLKK---REVQLLLANPGPIVIEKLHA 613
+ P + ++I +M+ V ID++G+ L D+ K+L K +V+ AN I E +
Sbjct: 398 NLKLPILNAIIFDMTAVRAIDSTGLRVLVDIKKTLDKFSGHKVEYHFAN---IQSEAIKK 228
Query: 614 SKLSDIIGEDK 624
+ + D E+K
Sbjct: 227 ALIEDGFHEEK 195
>BQ144272 similar to GP|13487715|gb| sulfate transporter 1 {Lycopersicon
esculentum}, partial (3%)
Length = 377
Score = 34.7 bits (78), Expect = 0.12
Identities = 15/27 (55%), Positives = 21/27 (77%)
Frame = +3
Query: 608 IEKLHASKLSDIIGEDKIFSSVDDAVA 634
I+KLH S ++ +GEDKIF +V +AVA
Sbjct: 3 IDKLHTSNFANFLGEDKIFLTVAEAVA 83
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.138 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,474,920
Number of Sequences: 36976
Number of extensions: 198924
Number of successful extensions: 1107
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 1093
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1098
length of query: 643
length of database: 9,014,727
effective HSP length: 102
effective length of query: 541
effective length of database: 5,243,175
effective search space: 2836557675
effective search space used: 2836557675
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0314.13


