

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
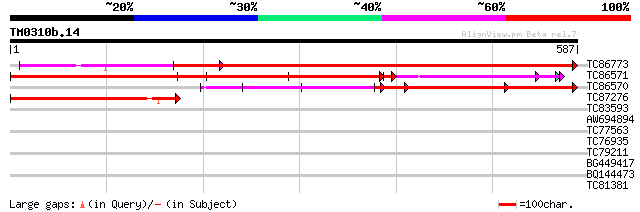
Query= TM0310b.14
(587 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86773 GP|11094367|gb|AAG29594.1 Ser/Thr specific protein phosp... 784 0.0
TC86571 homologue to GP|11094365|gb|AAG29593.1 Ser/Thr specific ... 709 0.0
TC86570 homologue to GP|11094365|gb|AAG29593.1 Ser/Thr specific ... 358 2e-99
TC87276 GP|11094367|gb|AAG29594.1 Ser/Thr specific protein phosp... 286 1e-77
TC83593 similar to PIR|B96671|B96671 similar to translational ac... 38 0.012
AW694894 similar to GP|12313690|db hypothetical protein {Oryza s... 32 0.52
TC77563 similar to GP|15450687|gb|AAK96615.1 AT5g59550/f2o15_210... 30 2.0
TC76935 similar to SP|P21747|EA92_VICFA Embryonic abundant prote... 29 5.8
TC79211 similar to GP|4204793|gb|AAD10836.1| P-glycoprotein {Sol... 29 5.8
BG449417 similar to GP|15810385|gb| unknown protein {Arabidopsis... 28 9.9
BQ144473 28 9.9
TC81381 similar to GP|10177135|dbj|BAB10425. gene_id:K2A18.28~pi... 28 9.9
>TC86773 GP|11094367|gb|AAG29594.1 Ser/Thr specific protein phosphatase 2A A
regulatory subunit beta isoform, partial (71%)
Length = 1626
Score = 784 bits (2024), Expect = 0.0
Identities = 399/418 (95%), Positives = 413/418 (98%)
Frame = +3
Query: 170 QLCQDDMPMVRRSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAAL 229
QLCQDDMPMVRRSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAAL
Sbjct: 3 QLCQDDMPMVRRSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAAL 182
Query: 230 GKLLEPQECVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLL 289
GKLLEPQ+CVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGP+ T+TELVPAYVRLL
Sbjct: 183 GKLLEPQDCVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPDSTKTELVPAYVRLL 362
Query: 290 RDNEAEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPV 349
RDNEAEVRIAAAGKVTKFSRILSPELAI+HILPCVKELS DSSQHVRSALASVIMGMAPV
Sbjct: 363 RDNEAEVRIAAAGKVTKFSRILSPELAIQHILPCVKELSTDSSQHVRSALASVIMGMAPV 542
Query: 350 LGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDR 409
LGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDR
Sbjct: 543 LGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDR 722
Query: 410 HWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGP 469
HWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAA N+KRLAEEFGP
Sbjct: 723 HWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAATNIKRLAEEFGP 902
Query: 470 DWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPN 529
+WAMQHIIPQVLDM+NDPHYLYRMTIL AISLLAPVLGSEIT++ LLPLVVNA+KDRVPN
Sbjct: 903 EWAMQHIIPQVLDMVNDPHYLYRMTILHAISLLAPVLGSEITTTNLLPLVVNAAKDRVPN 1082
Query: 530 IKFNVAKVLQSLIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 587
IKFNVAKVLQSLIPIV+ESVVE+T++PCLVELSEDPDVDVRFFASQALQSSDQVKMSS
Sbjct: 1083IKFNVAKVLQSLIPIVDESVVESTIKPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 1256
Score = 43.9 bits (102), Expect = 2e-04
Identities = 45/218 (20%), Positives = 90/218 (40%), Gaps = 6/218 (2%)
Frame = +3
Query: 11 IAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDDD-EVLLAMAE 69
+ + + LK+E +RLN I +L + + +G + + L+P + E +D V LA+ E
Sbjct: 573 LPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIE 752
Query: 70 ELGVFIPYVGGVEHANVLLPPLETLCTV----EETCVRDKSVESLCRIGAQMREQDLVEH 125
+IP + L LC + +RD + ++ R+ + + ++H
Sbjct: 753 ----YIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAATNIKRLAEEFGPEWAMQH 920
Query: 126 FIPLVKRLAAGEWFTARVSSCGLFHIAYP-SAPEAQKAELRAMYGQLCQDDMPMVRRSAA 184
IP V + + R++ + P E L + +D +P ++ + A
Sbjct: 921 IIPQVLDMVNDPHYLYRMTILHAISLLAPVLGSEITTTNLLPLVVNAAKDRVPNIKFNVA 1100
Query: 185 TNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLA 222
L V+ + ++S I +L++D VR A
Sbjct: 1101KVLQSLIPIVDESVVESTIKPCLVELSEDPDVDVRFFA 1214
>TC86571 homologue to GP|11094365|gb|AAG29593.1 Ser/Thr specific protein
phosphatase 2A A regulatory subunit alpha isoform,
partial (67%)
Length = 1490
Score = 709 bits (1830), Expect(2) = 0.0
Identities = 356/388 (91%), Positives = 376/388 (96%)
Frame = +2
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
M+M D+PLYPIAVLIDELKN+DIQLRLNSIRRLSTIARALGEERTRKELIPFL+ENNDDD
Sbjct: 287 MSMADEPLYPIAVLIDELKNDDIQLRLNSIRRLSTIARALGEERTRKELIPFLTENNDDD 466
Query: 61 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ 120
DEVLLAMAEELGVFIPYVGGVEHA+VLLPPLE C+VEETCVRDK+VESLCRIG+QMRE
Sbjct: 467 DEVLLAMAEELGVFIPYVGGVEHASVLLPPLEAFCSVEETCVRDKAVESLCRIGSQMRES 646
Query: 121 DLVEHFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVR 180
DLVE+FIPLVKRLAAGEWFTARVS+CGLFHIAYPSAPEA K ELR++Y QLCQDDMPMVR
Sbjct: 647 DLVEYFIPLVKRLAAGEWFTARVSACGLFHIAYPSAPEASKTELRSIYSQLCQDDMPMVR 826
Query: 181 RSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVA 240
RSAATNLGKFAATVE HLK+DIMS+FDDLTQDDQDSVRLLAVEGCAALGKLLEPQ+CVA
Sbjct: 827 RSAATNLGKFAATVEYTHLKADIMSIFDDLTQDDQDSVRLLAVEGCAALGKLLEPQDCVA 1006
Query: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA 300
HILPVIVNFSQDKSWRVRYMVANQLYELCEAV PEPT+ ELVPAYVRLLRDNEAEVRIAA
Sbjct: 1007HILPVIVNFSQDKSWRVRYMVANQLYELCEAVSPEPTKAELVPAYVRLLRDNEAEVRIAA 1186
Query: 301 AGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
AGKVTKF RILSP+LAI+HILPCVKELS+DSSQHVRSALASVIMGMAPVLGK+ATIEQLL
Sbjct: 1187AGKVTKFCRILSPDLAIQHILPCVKELSSDSSQHVRSALASVIMGMAPVLGKEATIEQLL 1366
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQV 388
PIFLSLLKDEFPDVRLNIISKLDQVNQV
Sbjct: 1367PIFLSLLKDEFPDVRLNIISKLDQVNQV 1450
Score = 111 bits (278), Expect = 7e-25
Identities = 90/376 (23%), Positives = 157/376 (40%)
Frame = +2
Query: 174 DDMPMVRRSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLL 233
DD V + A LG F V S ++ + ++ VR AVE +G +
Sbjct: 458 DDDDEVLLAMAEELGVFIPYVGGVEHASVLLPPLEAFCSVEETCVRDKAVESLCRIGSQM 637
Query: 234 EPQECVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNE 293
+ V + +P++ + + W + A L+ + PE ++TEL Y +L +D+
Sbjct: 638 RESDLVEYFIPLVKRLAAGE-WFTARVSACGLFHIAYPSAPEASKTELRSIYSQLCQDDM 814
Query: 294 AEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKD 353
VR +AA + KF+ + L AD
Sbjct: 815 PMVRRSAATNLGKFAATVE-----------YTHLKAD----------------------- 892
Query: 354 ATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRV 413
++ IF L +D+ VRL + + +++ +LP IV ++D+ WRV
Sbjct: 893 -----IMSIFDDLTQDDQDSVRLLAVEGCAALGKLLEPQDCVAHILPVIVNFSQDKSWRV 1057
Query: 414 RLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAM 473
R + + L + +L ++ L+D +R AAA V + PD A+
Sbjct: 1058RYMVANQLYELCEAVSPEPTKAELVPAYVRLLRDNEAEVRIAAAGKVTKFCRILSPDLAI 1237
Query: 474 QHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFN 533
QHI+P V ++ +D R + I +APVLG E T +LLP+ ++ KD P+++ N
Sbjct: 1238QHILPCVKELSSDSSQHVRSALASVIMGMAPVLGKEATIEQLLPIFLSLLKDEFPDVRLN 1417
Query: 534 VAKVLQSLIPIVEESV 549
+ L + + ES+
Sbjct: 1418IISKLDQVNQVYWESI 1465
Score = 63.2 bits (152), Expect = 3e-10
Identities = 73/367 (19%), Positives = 137/367 (36%)
Frame = +2
Query: 204 MSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHILPVIVNFSQDKSWRVRYMVAN 263
++V D ++D +RL ++ + + + L + ++P + + D V +A
Sbjct: 317 IAVLIDELKNDDIQLRLNSIRRLSTIARALGEERTRKELIPFLTENNDDDD-EVLLAMAE 493
Query: 264 QLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFSRILSPELAIEHILPC 323
+L VG + L+P E VR A + + + +E+ +P
Sbjct: 494 ELGVFIPYVGGVEHASVLLPPLEAFCSVEETCVRDKAVESLCRIGSQMRESDLVEYFIPL 673
Query: 324 VKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLD 383
VK L+A R + A + +A +A+ +L I+ L +D+ P VR + + L
Sbjct: 674 VKRLAAGEWFTARVS-ACGLFHIAYPSAPEASKTELRSIYSQLCQDDMPMVRRSAATNLG 850
Query: 384 QVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQ 443
+ A +EY L A + + FDD
Sbjct: 851 KF-----------------------------AATVEYTHLKADIMSI--FDDLT------ 919
Query: 444 WLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLA 503
+D S+R A L + P + HI+P +++ D + R + + L
Sbjct: 920 --QDDQDSVRLLAVEGCAALGKLLEPQDCVAHILPVIVNFSQDKSWRVRYMVANQLYELC 1093
Query: 504 PVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSLIPIVEESVVENTLRPCLVELSE 563
+ E T ++L+P V +D ++ A + I+ + + PC+ ELS
Sbjct: 1094EAVSPEPTKAELVPAYVRLLRDNEAEVRIAAAGKVTKFCRILSPDLAIQHILPCVKELSS 1273
Query: 564 DPDVDVR 570
D VR
Sbjct: 1274DSSQHVR 1294
Score = 61.6 bits (148), Expect = 8e-10
Identities = 59/286 (20%), Positives = 121/286 (41%)
Frame = +2
Query: 289 LRDNEAEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAP 348
L++++ ++R+ + +++ +R L E + ++P + E + D V A+A + P
Sbjct: 338 LKNDDIQLRLNSIRRLSTIARALGEERTRKELIPFLTE-NNDDDDEVLLAMAEELGVFIP 514
Query: 349 VLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAED 408
+G LLP + E VR + L ++ + L + +P + LA
Sbjct: 515 YVGGVEHASVLLPPLEAFCSVEETCVRDKAVESLCRIGSQMRESDLVEYFIPLVKRLAAG 694
Query: 409 RHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFG 468
+ R++ + A +L ++ Q +D + +R +AA N+ + A
Sbjct: 695 EWFTARVSACGLFHI-AYPSAPEASKTELRSIYSQLCQDDMPMVRRSAATNLGKFAATVE 871
Query: 469 PDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVP 528
I+ D+ D R+ ++ + L +L + + +LP++VN S+D+
Sbjct: 872 YTHLKADIMSIFDDLTQDDQDSVRLLAVEGCAALGKLLEPQDCVAHILPVIVNFSQDKSW 1051
Query: 529 NIKFNVAKVLQSLIPIVEESVVENTLRPCLVELSEDPDVDVRFFAS 574
+++ VA L L V + L P V L D + +VR A+
Sbjct: 1052RVRYMVANQLYELCEAVSPEPTKAELVPAYVRLLRDNEAEVRIAAA 1189
Score = 25.8 bits (55), Expect(2) = 0.0
Identities = 11/13 (84%), Positives = 13/13 (99%)
Frame = +1
Query: 388 VIGIDLLSQSLLP 400
++GIDLLSQSLLP
Sbjct: 1450 LLGIDLLSQSLLP 1488
>TC86570 homologue to GP|11094365|gb|AAG29593.1 Ser/Thr specific protein
phosphatase 2A A regulatory subunit alpha isoform,
partial (35%)
Length = 883
Score = 358 bits (920), Expect = 2e-99
Identities = 179/210 (85%), Positives = 198/210 (94%)
Frame = +2
Query: 378 IISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKL 437
IISKLDQVNQVIGIDL +SLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKL
Sbjct: 2 IISKLDQVNQVIGIDLYLKSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKL 181
Query: 438 GALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQVLDMINDPHYLYRMTILQ 497
GALCMQWL+DKV+SIR+AAANN+KRLAEEFGP+WAMQHIIPQVL+MI +PHYLYRMTIL+
Sbjct: 182 GALCMQWLQDKVHSIREAAANNLKRLAEEFGPEWAMQHIIPQVLEMITNPHYLYRMTILR 361
Query: 498 AISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSLIPIVEESVVENTLRPC 557
AISLLAPV+GSE+T SKLLP VV ASKDRVPNIKFNVAKVL+S+ PIV++SVVE T+RPC
Sbjct: 362 AISLLAPVMGSEVTCSKLLPAVVAASKDRVPNIKFNVAKVLESIFPIVDQSVVEKTIRPC 541
Query: 558 LVELSEDPDVDVRFFASQALQSSDQVKMSS 587
LVEL EDPDVDVRFF++QALQ+ D V MSS
Sbjct: 542 LVELGEDPDVDVRFFSNQALQAIDHVMMSS 631
Score = 92.0 bits (227), Expect = 6e-19
Identities = 49/173 (28%), Positives = 90/173 (51%)
Frame = +2
Query: 242 ILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAA 301
+LP IV ++D+ WRVR + + L +G +L ++ L+D +R AAA
Sbjct: 62 LLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLQDKVHSIREAAA 241
Query: 302 GKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLP 361
+ + + PE A++HI+P V E+ + R + I +APV+G + T +LLP
Sbjct: 242 NNLKRLAEEFGPEWAMQHIIPQVLEMITNPHYLYRMTILRAISLLAPVMGSEVTCSKLLP 421
Query: 362 IFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVR 414
++ KD P+++ N+ L+ + ++ ++ +++ P +VEL ED VR
Sbjct: 422 AVVAASKDRVPNIKFNVAKVLESIFPIVDQSVVEKTIRPCLVELGEDPDVDVR 580
Score = 62.0 bits (149), Expect = 6e-10
Identities = 49/215 (22%), Positives = 101/215 (46%)
Frame = +2
Query: 303 KVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPI 362
K+ + ++++ +L ++ +LP + EL+ D VR A+ I +A LG ++L +
Sbjct: 11 KLDQVNQVIGIDLYLKSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGAL 190
Query: 363 FLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIP 422
+ L+D+ +R + L ++ + G + Q ++P ++E+ + H+ R+ I+ I
Sbjct: 191 CMQWLQDKVHSIREAAANNLKRLAEEFGPEWAMQHIIPQVLEMITNPHYLYRMTILRAIS 370
Query: 423 LLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQVLD 482
LLA +G KL + KD+V +I+ A ++ + + I P +++
Sbjct: 371 LLAPVMGSEVTCSKLLPAVVAASKDRVPNIKFNVAKVLESIFPIVDQSVVEKTIRPCLVE 550
Query: 483 MINDPHYLYRMTILQAISLLAPVLGSEITSSKLLP 517
+ DP R QA+ + V+ S T + P
Sbjct: 551 LGEDPDVDVRFFSNQALQAIDHVMMSS*TIM*MCP 655
Score = 56.6 bits (135), Expect = 3e-08
Identities = 42/192 (21%), Positives = 83/192 (42%)
Frame = +2
Query: 198 HLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHILPVIVNFSQDKSWRV 257
+LKS ++ +L +D VRL +E L L + + + + QDK +
Sbjct: 50 YLKS-LLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLQDKVHSI 226
Query: 258 RYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFSRILSPELAI 317
R AN L L E GPE ++P + ++ + R+ ++ + ++ E+
Sbjct: 227 REAAANNLKRLAEEFGPEWAMQHIIPQVLEMITNPHYLYRMTILRAISLLAPVMGSEVTC 406
Query: 318 EHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLN 377
+LP V S D +++ +A V+ + P++ + + + P + L +D DVR
Sbjct: 407 SKLLPAVVAASKDRVPNIKFNVAKVLESIFPIVDQSVVEKTIRPCLVELGEDPDVDVRFF 586
Query: 378 IISKLDQVNQVI 389
L ++ V+
Sbjct: 587 SNQALQAIDHVM 622
>TC87276 GP|11094367|gb|AAG29594.1 Ser/Thr specific protein phosphatase 2A A
regulatory subunit beta isoform, partial (29%)
Length = 770
Score = 286 bits (733), Expect = 1e-77
Identities = 151/182 (82%), Positives = 159/182 (86%), Gaps = 5/182 (2%)
Frame = +2
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD
Sbjct: 236 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 415
Query: 61 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ 120
DEVLLA AEELGVF+PYVGGVEHA+VLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ
Sbjct: 416 DEVLLATAEELGVFVPYVGGVEHASVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ 595
Query: 121 DLVEHFIPLVKRLAAGEWFTARVSSCGLFHIA---YPSAPEAQKAELR--AMYGQLCQDD 175
DLVEHFIPLVKRLA+GEWFTA G H+A P+ ++ +L GQLCQDD
Sbjct: 596 DLVEHFIPLVKRLASGEWFTAP----GFLHVACFILPTLVHRRR*KLN*GPYMGQLCQDD 763
Query: 176 MP 177
MP
Sbjct: 764 MP 769
Score = 32.0 bits (71), Expect = 0.68
Identities = 28/125 (22%), Positives = 54/125 (42%), Gaps = 4/125 (3%)
Frame = +2
Query: 360 LPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIE 419
+ + + LK+E +RLN I +L + + +G + + L+P + E +D V LA E
Sbjct: 266 IAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDDD-EVLLATAE 442
Query: 420 ----YIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQH 475
++P + L LC + +RD + ++ R+ + ++H
Sbjct: 443 ELGVFVPYVGGVEHASVLLPPLETLCTV----EETCVRDKSVESLCRIGAQMREQDLVEH 610
Query: 476 IIPQV 480
IP V
Sbjct: 611 FIPLV 625
>TC83593 similar to PIR|B96671|B96671 similar to translational activator
[imported] - Arabidopsis thaliana, partial (8%)
Length = 675
Score = 37.7 bits (86), Expect = 0.012
Identities = 36/165 (21%), Positives = 77/165 (45%), Gaps = 6/165 (3%)
Frame = +3
Query: 355 TIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLP---AIVELAEDRH- 410
+++ +LPIFL L ++R L ++ +V G L + ++P ++ + DR
Sbjct: 120 SLQPILPIFLQGLISGSAELREQAALGLGELIEVAGEQSLKEVVIPITGPLIRIIGDRFP 299
Query: 411 WRVRLAIIEYIPLLASQLGVGF--FDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFG 468
W+V+ AI+ + ++ + G+ F +L ++ L+D +IR AA + L+ G
Sbjct: 300 WQVKSAILSTLTIMIRKGGISLKPFLPQLQTTFVKCLQDNTRTIRSGAAVALGMLS---G 470
Query: 469 PDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSS 513
+ + ++ +L + R IL A+ + G ++S+
Sbjct: 471 LNTRVDPLVSDLLSSLQGSDGGVREAILSALKGVLKHAGKNVSSA 605
>AW694894 similar to GP|12313690|db hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (15%)
Length = 665
Score = 32.3 bits (72), Expect = 0.52
Identities = 17/64 (26%), Positives = 30/64 (46%)
Frame = -3
Query: 468 GPDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRV 527
GP W +I + + P+ ++ ++ AP+ S +T++ LLPL K R
Sbjct: 567 GPAWV*IPLIATLFSVYTSPYLALSLSSTLIVTSCAPIQASRLTNTSLLPLGSATIKARD 388
Query: 528 PNIK 531
N+K
Sbjct: 387 RNLK 376
>TC77563 similar to GP|15450687|gb|AAK96615.1 AT5g59550/f2o15_210 {Arabidopsis
thaliana}, partial (39%)
Length = 1468
Score = 30.4 bits (67), Expect = 2.0
Identities = 25/89 (28%), Positives = 46/89 (51%), Gaps = 6/89 (6%)
Frame = -2
Query: 279 TELVPAYVRLLRDNEAE---VRIAAAGKVTKFSRILS-PELAIEHILPCVKELSADSSQH 334
T+++ + LLR+NE E ++ KF +IL P L++ +LP VKE+ +SS
Sbjct: 1107 TQVLLLLLELLRNNEPEKLRFKLTFPKNDKKFRKILLIPRLSLTLLLPTVKEILLESSFE 928
Query: 335 VRSALASVIMG--MAPVLGKDATIEQLLP 361
+ +++ +G ++P L A + P
Sbjct: 927 LLPSISV*TIGN*ISPALRPPANLPTANP 841
>TC76935 similar to SP|P21747|EA92_VICFA Embryonic abundant protein USP92
precursor. [Broad bean] {Vicia faba}, partial (40%)
Length = 1840
Score = 28.9 bits (63), Expect = 5.8
Identities = 12/43 (27%), Positives = 25/43 (57%)
Frame = +1
Query: 502 LAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSLIPI 544
+ P++ S+ T SK L + + ++ PN+ ++V KV +P+
Sbjct: 1387 MVPLVASDGTKSKALTICHHDTRGMDPNVLYDVLKVKPGTVPV 1515
>TC79211 similar to GP|4204793|gb|AAD10836.1| P-glycoprotein {Solanum
tuberosum}, partial (36%)
Length = 1667
Score = 28.9 bits (63), Expect = 5.8
Identities = 24/80 (30%), Positives = 38/80 (47%), Gaps = 1/80 (1%)
Frame = -2
Query: 64 LLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQDLV 123
L+A ++L +F P G L PLE + ++ E+ R+ ++ REQ L
Sbjct: 1279 LIAYVQQLWLFFPLKRG-------LAPLELISLIQHQVHLLPRQEA*ARLSSKKREQLLF 1121
Query: 124 EHFIPL-VKRLAAGEWFTAR 142
F+PL V+RL + F R
Sbjct: 1120 FVFVPLRVERLFPLQMFCIR 1061
>BG449417 similar to GP|15810385|gb| unknown protein {Arabidopsis thaliana},
partial (13%)
Length = 630
Score = 28.1 bits (61), Expect = 9.9
Identities = 16/63 (25%), Positives = 32/63 (50%), Gaps = 5/63 (7%)
Frame = +2
Query: 9 YPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTR-----KELIPFLSENNDDDDEV 63
+P DEL+ +D+ L +I S+ +++GE TR ++++ +S+ + V
Sbjct: 41 WPTKKTTDELEEQDLDLASQTISAQSSSGKSMGESNTRTTSRTEQILSSVSDEETKHEGV 220
Query: 64 LLA 66
L A
Sbjct: 221 LEA 229
>BQ144473
Length = 873
Score = 28.1 bits (61), Expect = 9.9
Identities = 12/30 (40%), Positives = 17/30 (56%)
Frame = +3
Query: 319 HILPCVKELSADSSQHVRSALASVIMGMAP 348
H LPC+ +L H+ SAL ++ MAP
Sbjct: 222 HHLPCLSDLLPKPHPHIHSALQILLDSMAP 311
>TC81381 similar to GP|10177135|dbj|BAB10425.
gene_id:K2A18.28~pir||T10240~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (34%)
Length = 965
Score = 28.1 bits (61), Expect = 9.9
Identities = 14/32 (43%), Positives = 19/32 (58%)
Frame = +3
Query: 277 TRTELVPAYVRLLRDNEAEVRIAAAGKVTKFS 308
T T ++ VRLL + EAEV AA + KF+
Sbjct: 366 TETRIIGPLVRLLDEREAEVSKEAADSLAKFA 461
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.136 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,820,899
Number of Sequences: 36976
Number of extensions: 203151
Number of successful extensions: 908
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 876
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 896
length of query: 587
length of database: 9,014,727
effective HSP length: 101
effective length of query: 486
effective length of database: 5,280,151
effective search space: 2566153386
effective search space used: 2566153386
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0310b.14


