

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
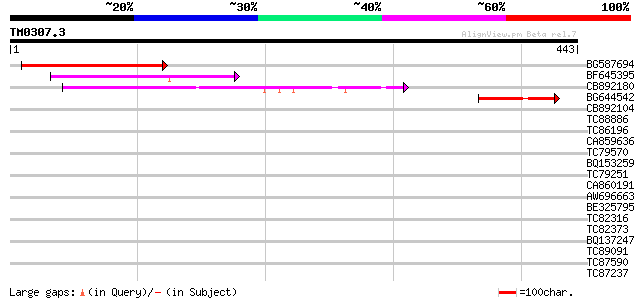
Query= TM0307.3
(443 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG587694 similar to GP|20260480|gb unknown protein {Arabidopsis ... 134 5e-32
BF645395 similar to GP|15808946|gb| auxin-regulated protein {Lyc... 106 2e-23
CB892180 similar to PIR|D84681|D84 hypothetical protein At2g2815... 104 8e-23
BG644542 similar to PIR|D84681|D846 hypothetical protein At2g281... 50 2e-06
CB892104 38 0.007
TC88886 similar to PIR|T49136|T49136 protein kinase-like protein... 35 0.044
TC86196 similar to GP|22652125|gb|AAN03626.1 BEL1-related homeot... 33 0.17
CA859636 33 0.17
TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (AR... 33 0.22
BQ153259 homologue to SP|P37617|ATZN_ Lead cadmium zinc and me... 33 0.29
TC79251 similar to GP|21555541|gb|AAM63881.1 unknown {Arabidopsi... 32 0.38
CA860191 homologue to GP|19697342|gb hypothetical protein {Dicty... 31 0.84
AW696663 weakly similar to GP|17827193|dbj ORF1 {TT virus}, part... 31 0.84
BE325795 similar to GP|22136848|gb| unknown protein {Arabidopsis... 31 1.1
TC82316 similar to GP|19807751|gb|AAL99323.1 hypothetical protei... 30 1.4
TC82373 weakly similar to OMNI|NT01MC0976 conserved hypothetical... 30 1.4
BQ137247 30 1.9
TC89091 similar to GP|10176779|dbj|BAB09893. pollen specific pro... 30 1.9
TC87590 weakly similar to PIR|G96522|G96522 F11A17.16 [imported]... 30 1.9
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 30 2.4
>BG587694 similar to GP|20260480|gb unknown protein {Arabidopsis thaliana},
partial (36%)
Length = 754
Score = 134 bits (338), Expect = 5e-32
Identities = 67/115 (58%), Positives = 83/115 (71%), Gaps = 1/115 (0%)
Frame = +1
Query: 10 RCSSRDTSPDRAKICRKVI-KPARKVQVVYYLFRNGLLEHPHFMELTLLPNQPLRLKDVF 68
R +R+TSP+R K+ + K RKV VVYYL RNG LEHPHFME+ L L LKDV
Sbjct: 79 RPGTRETSPERTKVWTEPKPKTPRKVSVVYYLSRNGHLEHPHFMEVPLSSPHGLYLKDVI 258
Query: 69 DRLMVLRGSGMPLQYSWSSKRNYKSGYVWFDLALKDVIYPSEGGEYVLKGSELVE 123
RL +LRG GMP YSWSSKR+YK+G+VW DL D IYP++G +Y+LKGSE++E
Sbjct: 259 IRLNLLRGKGMPTMYSWSSKRSYKNGFVWHDLTESDFIYPTQGHDYILKGSEILE 423
>BF645395 similar to GP|15808946|gb| auxin-regulated protein {Lycopersicon
esculentum}, partial (19%)
Length = 658
Score = 106 bits (264), Expect = 2e-23
Identities = 63/162 (38%), Positives = 87/162 (52%), Gaps = 15/162 (9%)
Frame = +1
Query: 33 KVQVVYYLFRNGLLEHPHFMELTLLPNQ-PLRLKDVFDRLMVLRGSGMPLQYSWSSKRNY 91
KV V+YYL RNG LEHPH M +++ + L L+DV +RL LRG G+ YSWS+KR+Y
Sbjct: 58 KVPVIYYLSRNGQLEHPHLMYVSISSSHGTLCLQDVINRLSFLRGQGIANMYSWSTKRSY 237
Query: 92 KSGYVWFDLALKDVIYPSEGGEYVLKGSELVE--------------GCSERFQQLNVSNK 137
++G+VW DL+ D+IYPS EYVLKG+ L+E S+ + N S+
Sbjct: 238 RNGFVWQDLSENDLIYPSSSHEYVLKGTLLIEEPSSFRSYETILSMPSSKSSNETNNSSS 417
Query: 138 NGIHEPPETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEEK 179
P T G K+ ++ AE+ Q EEK
Sbjct: 418 MDADSPSSTAKGSRRDYKLYKATTYREFAEKATNASTQTEEK 543
>CB892180 similar to PIR|D84681|D84 hypothetical protein At2g28150 [imported]
- Arabidopsis thaliana, partial (29%)
Length = 872
Score = 104 bits (259), Expect = 8e-23
Identities = 91/294 (30%), Positives = 134/294 (44%), Gaps = 24/294 (8%)
Frame = +2
Query: 42 RNGLLEHPHFMELTLLPNQPLRLKDVFDRLMVLRGSGMPLQYSWSSKRNYKSGYVWFDLA 101
R+ LEHPHFME+ + L L+DV +L LRG GM YSWS KR+YK+GYVW DL+
Sbjct: 5 RDRQLEHPHFMEVPISSPDGLFLRDVIHKLDALRGRGMADLYSWSCKRSYKNGYVWHDLS 184
Query: 102 LKDVIYPSEGGEYVLKGSEL-VEGCSERFQQLNVSNKNGIHEPPETNYGYNAKSKVLSNR 160
D+I P+ G EYVLKGSEL E S+RF ++ + PE + +V S+
Sbjct: 185 EDDLILPAHGNEYVLKGSELFCESNSDRFSPISNVKLQSLKRLPEP-VSCRSHDEVSSSS 361
Query: 161 QQQKEAEEYEEYEEQEEEKEFT-SSTTTPHSRCSRGVS------------TEELGEDEDR 207
E E E++ ++ T SS +P S + S T+ L + +
Sbjct: 362 SSMNEREGRNSQEDEISPRQHTGSSDISPESSDGKSDSQSLPLTEYKIYKTDRLADASTQ 541
Query: 208 HE--ALQNNTKTEKV----VVTNNVSSRGNVTNVPPPMKNHSAAAASLAEKLNRRVVVPV 261
E A ++ ++T K V T + SS ++ H + + VV P
Sbjct: 542 TEECAAKHQSQTHKTCTRGVSTEDRSSESECNDICETQAGHVKDGSEICSD----VVSPP 709
Query: 262 ---QSKQRFGGELGT-ESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVS 311
S GG+ T ES A SR ++ L G + A T+ +P ++
Sbjct: 710 FSNSSPSSSGGKTETLESLIRADASRMNSFRIL---EEQGIQMPASTRLKPSIA 862
>BG644542 similar to PIR|D84681|D846 hypothetical protein At2g28150
[imported] - Arabidopsis thaliana, partial (5%)
Length = 781
Score = 49.7 bits (117), Expect = 2e-06
Identities = 27/64 (42%), Positives = 39/64 (60%), Gaps = 1/64 (1%)
Frame = -3
Query: 367 EEKEYFSGSLVEME-SMKANHPVLKRSNSYNEERRSKLGVEEVKLKKEMEEENRERKGGV 425
++KEYFSGSLVE + +M+ H VLKRS+S+N ER S + + KE++ +N
Sbjct: 641 DDKEYFSGSLVESKMAMEDGHNVLKRSSSFNAERTS---AQTYRTSKELKSQNMVESSSG 471
Query: 426 KEKC 429
KC
Sbjct: 470 NSKC 459
>CB892104
Length = 560
Score = 38.1 bits (87), Expect = 0.007
Identities = 35/156 (22%), Positives = 66/156 (41%), Gaps = 12/156 (7%)
Frame = +1
Query: 120 ELVEGCSERFQQLNVSNKNGIHEPPETNYGY----NAKSKV---LSNRQQQKEAEEYE-- 170
+L E C ++ L VSN HE P+ N+K + + + K EE E
Sbjct: 76 DLNENCHSSYECLIVSNDTQSHENPQEKEKLDDDDNSKEMMKLKIVHESSTKSGEETETS 255
Query: 171 --EYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEALQNNTKTEKVVVTNNVSS 228
+ E+ E+ +++T + S S+ + +E D++ H ++ + T+ +
Sbjct: 256 SFKQEQFEKVEDYTGGGSDVESSRSKEIKRDESSGDDELHPSI-SPTRLNSANKSKTDEE 432
Query: 229 RGNVTNVPPPM-KNHSAAAASLAEKLNRRVVVPVQS 263
++ PPP+ KN + S KLN + +S
Sbjct: 433 TKHLQQSPPPLVKNLQIPSYSYTNKLNPNTSMAAES 540
>TC88886 similar to PIR|T49136|T49136 protein kinase-like protein -
Arabidopsis thaliana, partial (14%)
Length = 1351
Score = 35.4 bits (80), Expect = 0.044
Identities = 61/304 (20%), Positives = 111/304 (36%), Gaps = 26/304 (8%)
Frame = +2
Query: 166 AEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGE------------DEDR---HEA 210
AEE E + E T S PHS EE+G D+D+ E
Sbjct: 74 AEEIEGLRDTSNEMALTKSLKHPHSVSGEKSICEEVGSVNKLNNRPETVTDDDKFTVKER 253
Query: 211 LQNNTKTEKVVVTNNVSS------RGNVTNVPP---PMKNHSAAAASLAEKLNRRVVVPV 261
L ++T V+ + SS +G VT P P +H AA + R V
Sbjct: 254 LSLVSETAPVITSTKASSQKVLQEKGTVTQNPAQERPDTSHLPAAFDDVIHVIRHSSYRV 433
Query: 262 QSKQRF--GGELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASK 319
S Q E+G ++ + + I ++ P+ ++ ++ + S
Sbjct: 434 GSDQPVMESVEMGVQNVD--VGKFINVMRDDIEMRNTNPQMTLKSSNSSEAETLKSNISD 607
Query: 320 QRQSVDRRDQKAFMMSCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEM 379
Q + R + + + +A ++ + +SENP+ + + E + S M
Sbjct: 608 QLEM--RNTSTSTPPTLKSSSFSDAASLKSSSISENPQDVSNTVSLVSESDSSEHSKCNM 781
Query: 380 ESMKANHPVLKRSNSYNEERRSKLGVEEVKLKKEMEEENRERKGGVKEKCIPIKKSSKES 439
+ K P + + + +R++ E ++L E+ + NR + V K K S
Sbjct: 782 PTTKDKPPEKEILDVKSFRQRAEALEELLELSAELLQHNRLEELQVVLKPFGKDKVSPRE 961
Query: 440 SRRW 443
+ W
Sbjct: 962 TAIW 973
>TC86196 similar to GP|22652125|gb|AAN03626.1 BEL1-related homeotic protein 29
{Solanum tuberosum}, partial (53%)
Length = 2227
Score = 33.5 bits (75), Expect = 0.17
Identities = 21/115 (18%), Positives = 54/115 (46%), Gaps = 5/115 (4%)
Frame = +1
Query: 311 SSVGTTASKQRQSVDRRDQKAFMMSCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKE 370
SS+ T+S+++ + K + + + ++ + ++R +P ++GN++N
Sbjct: 1552 SSMKITSSQEKALTSETEPKGYNSTQDISTSQDTPLISVSRPQTSP--LVGNVRNNSGFS 1725
Query: 371 YFSGSLVE-----MESMKANHPVLKRSNSYNEERRSKLGVEEVKLKKEMEEENRE 420
+ S +E NH ++ NS+ + + ++ EE+ +K + +NR+
Sbjct: 1726 FIGSSELEGIPQGSPKKPRNHELMHNPNSFIDVKHNEANNEELSMKFGDDRQNRD 1890
>CA859636
Length = 593
Score = 33.5 bits (75), Expect = 0.17
Identities = 30/118 (25%), Positives = 50/118 (41%), Gaps = 12/118 (10%)
Frame = +3
Query: 323 SVDRRDQKAFMMSCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSG---SLVEM 379
++D+ DQ + + C EE EEA+ M RV+ L + K Y SG L ++
Sbjct: 117 TMDQSDQPSEIQGCPEESPEEAL-MASFRVAAESVAQLYKTALNQTKRYDSGYEQCLRDL 293
Query: 380 ESMKANHPVLKRSNSYNEERRS---------KLGVEEVKLKKEMEEENRERKGGVKEK 428
A+HP ++RS + R K + + K + NRE V+++
Sbjct: 294 MEFVASHPSVQRSQQTGGDGRDAFISVEDLCKFAQSKTEQLKSISRRNREYIMNVRQQ 467
>TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (15%)
Length = 1143
Score = 33.1 bits (74), Expect = 0.22
Identities = 17/73 (23%), Positives = 39/73 (53%)
Frame = +2
Query: 126 SERFQQLNVSNKNGIHEPPETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEEKEFTSST 185
+++FQQL H+ TN Y+ + + +QQQ++ ++ +++++Q++++
Sbjct: 512 TQQFQQLLQ------HQHSFTNQNYHMQQQ--QQQQQQQQQQQQQQHQQQQQQQSQQQQQ 667
Query: 186 TTPHSRCSRGVST 198
H + S VST
Sbjct: 668 VVDHQQMSNAVST 706
>BQ153259 homologue to SP|P37617|ATZN_ Lead cadmium zinc and mercury
transporting ATPase (EC 3.6.3.3) (EC 3.6.3.5).
{Escherichia coli}, partial (8%)
Length = 343
Score = 32.7 bits (73), Expect = 0.29
Identities = 18/31 (58%), Positives = 19/31 (61%), Gaps = 1/31 (3%)
Frame = -2
Query: 269 GELGTESATSAAPS-RYSALLQLIACGSSGP 298
GELG E A RYSALLQL+ACG P
Sbjct: 180 GELGLEFKAGAIYCVRYSALLQLLACGKCSP 88
>TC79251 similar to GP|21555541|gb|AAM63881.1 unknown {Arabidopsis
thaliana}, partial (56%)
Length = 1253
Score = 32.3 bits (72), Expect = 0.38
Identities = 18/39 (46%), Positives = 23/39 (58%), Gaps = 4/39 (10%)
Frame = +2
Query: 184 STTTPHSRCSRG----VSTEELGEDEDRHEALQNNTKTE 218
STTT HSRCSRG + T EDE+ + +NT+ E
Sbjct: 2 STTTLHSRCSRGESINIRTRRNREDENTVGHIDSNTQIE 118
>CA860191 homologue to GP|19697342|gb hypothetical protein {Dictyostelium
discoideum}, partial (1%)
Length = 504
Score = 31.2 bits (69), Expect = 0.84
Identities = 19/54 (35%), Positives = 27/54 (49%)
Frame = +2
Query: 155 KVLSNRQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRH 208
K+ Q + +E EE E++EEE EF TTT +S + S +ED H
Sbjct: 212 KMQQGLDQTLQNDEEEEEEDEEEEDEF---TTTFNSNNNNNNSNNNNNNNEDNH 364
>AW696663 weakly similar to GP|17827193|dbj ORF1 {TT virus}, partial (2%)
Length = 660
Score = 31.2 bits (69), Expect = 0.84
Identities = 14/22 (63%), Positives = 18/22 (81%)
Frame = -1
Query: 159 NRQQQKEAEEYEEYEEQEEEKE 180
++QQQ E EE EE EE+EEE+E
Sbjct: 177 HQQQQLEEEEEEEEEEEEEERE 112
>BE325795 similar to GP|22136848|gb| unknown protein {Arabidopsis thaliana},
partial (6%)
Length = 676
Score = 30.8 bits (68), Expect = 1.1
Identities = 28/100 (28%), Positives = 47/100 (47%), Gaps = 13/100 (13%)
Frame = +2
Query: 155 KVLSN--RQQQKEAEEYEEYEEQEEEKEFTSSTTT----PHSRCSRGVSTEELGEDEDRH 208
K LSN R+ EAE+ EEYE+ ++K ++ T P++ S T+ + + +
Sbjct: 308 KQLSNISRRTLLEAEQEEEYEQPLKKKSSSTKNQTKLIKPNNNLSSKNQTKTI-KSNTNN 484
Query: 209 EALQNNTK-------TEKVVVTNNVSSRGNVTNVPPPMKN 241
+ +N TK T K +T+ +S N T P +N
Sbjct: 485 LSSKNQTKPSKDSKLTSKTTLTDTTTSLKNSTPQPSKQRN 604
>TC82316 similar to GP|19807751|gb|AAL99323.1 hypothetical protein
{Dictyostelium discoideum}, partial (1%)
Length = 632
Score = 30.4 bits (67), Expect = 1.4
Identities = 38/196 (19%), Positives = 79/196 (39%), Gaps = 24/196 (12%)
Frame = +1
Query: 157 LSNRQQQKEAEEYEEYEEQEEEK--EFTSSTTTPHSRCSRGVSTEELGEDEDRH------ 208
+ N + +EY++YEE+E + +ST +P + +D+D +
Sbjct: 49 VGNEESTLLQDEYDDYEEEETLSLCDLPNSTISPQRGDFSNYGKDHHDDDDDDNLFEFFS 228
Query: 209 EALQNNTKTEKVV-------------VTNNVSSRGNVTNVPPPMKNHSAAAASLAEKLNR 255
E +T + ++ V +N + T+ MK+ + + A+L K N
Sbjct: 229 EEFNTSTIHDNIIFCGKLIPFKDHQYVPHNQKNCAKPTSNSKAMKSSNGSIANLKSKRNE 408
Query: 256 RVVVPVQSKQRFGGE---LGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSS 312
V + + F G+ +G + + +P++ L + SS + +E ++S
Sbjct: 409 EEVKGSVNVKSFAGDYTSMGGKVSLVRSPTKSRWFLFMFGMSSS----SRMSSKEMQLSD 576
Query: 313 VGTTASKQRQSVDRRD 328
+ + RQS RR+
Sbjct: 577 I-----RNRQSRSRRE 609
>TC82373 weakly similar to OMNI|NT01MC0976 conserved hypothetical protein
{Magnetococcus sp. MC-1}, partial (11%)
Length = 787
Score = 30.4 bits (67), Expect = 1.4
Identities = 20/68 (29%), Positives = 31/68 (45%), Gaps = 1/68 (1%)
Frame = +3
Query: 162 QQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDE-DRHEALQNNTKTEKV 220
Q+ + E+ +E +Q EEK S GV G+D+ R + + KT+ V
Sbjct: 267 QKSQREKEDEIRKQLEEKAMERQRRIAERSASSGVGRTVPGKDQIARISSKTDKNKTQTV 446
Query: 221 VVTNNVSS 228
TN +SS
Sbjct: 447 KETNRISS 470
>BQ137247
Length = 998
Score = 30.0 bits (66), Expect = 1.9
Identities = 44/220 (20%), Positives = 83/220 (37%), Gaps = 18/220 (8%)
Frame = +2
Query: 158 SNRQQQKEAEE------------YEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDE 205
+NRQ+ +E ++ ++++ + K S+TT H R T D
Sbjct: 251 TNRQEWREKKKKTNEQDRTRGRGHDKHTTTDTHKHHQSTTTPAHIRDDNTRQTTLRRADS 430
Query: 206 DRHEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNHSAAAASLAEKLNRRVVVPVQSKQ 265
+ + K + V TN+ S + +P + A+A L E +Q +
Sbjct: 431 ESRDT--QGAKARRYVTTNSP*SENMILQIPKA*QESGASARRLDE---------IQDDK 577
Query: 266 RFGGELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASKQRQ--- 322
+ ++G + P+ +L K R + E R + G T K R+
Sbjct: 578 KVRAKVG------SGPTANDQHRELTVRKRCDKHTK*RKKAEERGAREGETHEKSRRNDI 739
Query: 323 ---SVDRRDQKAFMMSCEEEEEEEAVAMMMNRVSENPRLM 359
+ D+ +K ++ +E ++EEA R E+ R M
Sbjct: 740 EVATYDKASEK--KITVQETQQEEA----STRAEESRRTM 841
>TC89091 similar to GP|10176779|dbj|BAB09893. pollen specific protein SF21
{Arabidopsis thaliana}, partial (42%)
Length = 794
Score = 30.0 bits (66), Expect = 1.9
Identities = 19/45 (42%), Positives = 24/45 (53%)
Frame = +3
Query: 251 EKLNRRVVVPVQSKQRFGGELGTESATSAAPSRYSALLQLIACGS 295
EKLN R ++ V F E TS RYSAL+++ ACGS
Sbjct: 123 EKLNCRTLIFVGDSSPFHSE--AIHMTSKLDRRYSALVEVQACGS 251
>TC87590 weakly similar to PIR|G96522|G96522 F11A17.16 [imported] -
Arabidopsis thaliana, partial (45%)
Length = 2425
Score = 30.0 bits (66), Expect = 1.9
Identities = 53/253 (20%), Positives = 106/253 (40%), Gaps = 3/253 (1%)
Frame = +2
Query: 171 EYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEALQNNTKTEKVVVTNNVS--S 228
++ +Q++ + S T P +R S++ + + KT +V N VS S
Sbjct: 320 KHHQQQQHSDNKSLQTVPQTRLRVRASSKA-----------KESPKTPPEIV-NRVSTIS 463
Query: 229 RGNVTNVPPPMKNHSAAAASL-AEKLNRRVVVPVQSKQRFGGELGTESATSAAPSRYSAL 287
+VPP MKN+S A S+ K+ + + V+S + E AP R +
Sbjct: 464 STRAKSVPPDMKNNSKAKRSIFMNKVVKSIEEEVESSHKGSKEGEVAKVVVVAPPRRRRI 643
Query: 288 LQLIACGSSGPEFKARTQQEPRVSSVGTTASKQRQSVDRRDQKAFMMSCEEEEEEEAVAM 347
+ P+ K + + ++ V K QS ++ ++E + +
Sbjct: 644 EE------DDPDVKEKKELLEKL-EVSENLIKSLQS---------EINALKDELNQVKGL 775
Query: 348 MMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKANHPVLKRSNSYNEERRSKLGVEE 407
++ S+N +L NL +AE K G+ S + P+ +R + ++ + K+ ++
Sbjct: 776 NIDLESQNIKLN-QNLASAEAKIVAFGT---SSSTRKKEPIGERQSPKFKDIQ-KIIADK 940
Query: 408 VKLKKEMEEENRE 420
+++ K +E N E
Sbjct: 941 LEMSKVKKEANPE 979
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 29.6 bits (65), Expect = 2.4
Identities = 25/90 (27%), Positives = 43/90 (47%), Gaps = 9/90 (10%)
Frame = +1
Query: 362 NLQNAEEKEYFSGS-----LVEMESMKANHPVLKRSNSYNEERR----SKLGVEEVKLKK 412
+L+N E KE S L ES + ++S+ NEE + SK EE+K +
Sbjct: 13 HLENEESKETGESSEEKSHLENEESKETGESSEEKSHLENEENKDEEKSKQENEEIKDGE 192
Query: 413 EMEEENRERKGGVKEKCIPIKKSSKESSRR 442
++++EN E K K + + +E S++
Sbjct: 193 KIQQENEENKDEEKSQQENEENKDEEKSQQ 282
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.310 0.126 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,333,439
Number of Sequences: 36976
Number of extensions: 152912
Number of successful extensions: 845
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 818
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 837
length of query: 443
length of database: 9,014,727
effective HSP length: 99
effective length of query: 344
effective length of database: 5,354,103
effective search space: 1841811432
effective search space used: 1841811432
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0307.3


