

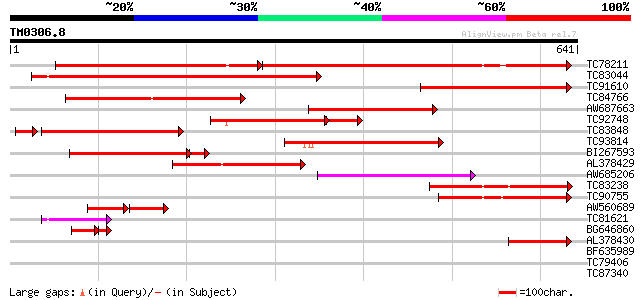
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0306.8
(641 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78211 weakly similar to GP|15624010|dbj|BAB68064. sulfate tran... 272 e-134
TC83044 similar to PIR|T48901|T48901 sulfate transporter ATST1 [... 305 3e-83
TC91610 weakly similar to PIR|T50022|T50022 sulfate transporter ... 281 5e-76
TC84766 similar to SP|P53392|SUT2_STYHA High affinity sulphate t... 240 1e-63
AW687663 homologue to GP|13487717|gb sulfate transporter 2 {Lyco... 199 3e-51
TC92748 similar to SP|P53393|SUT3_STYHA Low affinity sulphate tr... 165 4e-48
TC83848 similar to SP|P53393|SUT3_STYHA Low affinity sulphate tr... 169 2e-45
TC93814 similar to PIR|B86365|B86365 probable sulphate transport... 179 2e-45
BI267593 similar to PIR|B86365|B86 probable sulphate transporter... 163 2e-42
AL378429 similar to GP|9294446|dbj sulfate transporter {Arabidop... 148 6e-36
AW685206 similar to GP|9955547|emb sulfate transporter {Arabidop... 132 3e-31
TC83238 weakly similar to PIR|T49069|T49069 sulfate transporter ... 124 1e-28
TC90755 similar to SP|P53391|SUT1_STYHA High affinity sulphate t... 117 2e-26
AW560689 similar to GP|9294446|dbj| sulfate transporter {Arabido... 64 2e-16
TC81621 similar to PIR|T49069|T49069 sulfate transporter (ATST1)... 62 9e-10
BG646860 similar to GP|13487717|gb| sulfate transporter 2 {Lycop... 46 5e-08
AL378430 similar to GP|9294446|dbj| sulfate transporter {Arabido... 51 1e-06
BF635989 weakly similar to GP|9294446|dbj| sulfate transporter {... 39 0.005
TC79406 weakly similar to PIR|T04879|T04879 hypothetical protein... 32 0.75
TC87340 similar to GP|15215816|gb|AAK91453.1 At2g26990/T20P8.4 {... 31 1.7
>TC78211 weakly similar to GP|15624010|dbj|BAB68064. sulfate transporter-like
protein {Oryza sativa (japonica cultivar-group)}, partial
(66%)
Length = 2090
Score = 272 bits (696), Expect(2) = e-134
Identities = 143/351 (40%), Positives = 225/351 (63%), Gaps = 1/351 (0%)
Frame = +2
Query: 286 LSTLIVFLTRADKSGVKIVKHVKGGLNPSSLHELDFNNPHVGEAAKIGLVVAVVALTESI 345
L + +L ++ + G++IV H+ GLNP S+ L F+ ++ K GL+ V++L E I
Sbjct: 875 LGGVFTYLVKSTQHGIQIVGHLDKGLNPISIQFLTFDRRYLSTVMKAGLISGVLSLAEGI 1054
Query: 346 AVGRSFASIKGYQLDGNKEMMSIGFSNIIGSLTSCYVATGSFSRTAVNYAAGCETLISNI 405
A+GRSF+ DGNKEM++ G N++GS TSCY+ +G FS+TAVNY AGC++ ++N+
Sbjct: 1055 AIGRSFSVTANTPHDGNKEMIAFGLMNLVGSFTSCYLTSGPFSKTAVNYNAGCKSAMTNV 1234
Query: 406 VMAITVLISLQFFTKLLYYTPTAIIASVILSALPGLIDIPEACKIWKVDKLDFLACAGAF 465
V A+ + ++LQ L TP ++++I+SA+ GLI+ EA ++KVDK DF+ C AF
Sbjct: 1235 VQAVIMALTLQLLAPLFSNTPLVALSAIIVSAMLGLINYTEAIHLFKVDKFDFIICMSAF 1414
Query: 466 FGVLFASVEIGLLAAVMISFLKIILISIRPDTEALGKLPGTSLFCDVYQYPMAVQIPGVV 525
GV F S++IGL+ +V + L+ +L RP LGKLP + L+ DV QY A IPGV+
Sbjct: 1415 LGVAFLSMDIGLMLSVGLGVLRGLLYLARPPACKLGKLPDSGLYRDVEQYSNASTIPGVL 1594
Query: 526 VIRVKSALLCFANANFVRERIMKWVTQEESKDDKGNSTNTIQLVILETSNLVDIDTSGIA 585
+I+V S + F+N+ +++ERI++++ E+S +S + ++ VIL + + IDT+ I
Sbjct: 1595 IIQVGSPIY-FSNSTYLKERILRYIKSEQS-----SSGDMVEHVILVLTAVSSIDTTAIE 1756
Query: 586 SLEEMQKVLISNGKQLAIANPRWQVIHKLKVSNFVSKIGGR-IYLTVEEAI 635
L E QK+L G Q+A+ NPR +V+ KL S FV K+G YL +E+A+
Sbjct: 1757 GLLETQKILEMKGIQMALVNPRLEVMEKLIASKFVEKVGKESFYLNLEDAV 1909
Score = 226 bits (575), Expect(2) = e-134
Identities = 109/235 (46%), Positives = 156/235 (66%), Gaps = 1/235 (0%)
Frame = +1
Query: 53 KDQPCNTLMSFLQGIFPILSWGRNYTAAKFRKDILAGLTIASLCIPQSIGYATLAHLDPQ 112
+++ L+ +Q PI W NY+ F D++AGLTIASL IPQ I YA LA+L P
Sbjct: 178 EEKKSRRLIKGVQYFIPIFEWLPNYSLRLFFSDLIAGLTIASLAIPQGISYAKLANLPPL 357
Query: 113 YGLYTSVVPPLIYAVMGTSREVAVGPVAVVSLLLFSMVQKLVDPAVDPVAYTKLVFLTTL 172
GLY+S VPPL+YAV G+SR +AVG +A SLL+ S+V + DP +P Y L+F TT
Sbjct: 358 IGLYSSFVPPLVYAVFGSSRHMAVGTIAAASLLIASIVSTVADPIAEPTLYLHLIFTTTF 537
Query: 173 FAGIFQTAFGLFRLGFLVDFLSHAAIVGFVAGAAIVIGLQQFKGLLGITHFTTKTDIISV 232
G+FQ G FRLG LVDF SH+ I GF+ G A+++ LQQFKG+ G+ HF+TKT++++V
Sbjct: 538 ITGVFQACLGFFRLGILVDFFSHSTITGFMGGTAVILILQQFKGIFGMKHFSTKTNVVAV 717
Query: 233 MKAVWEALHN-PWQPRNFILGSSFLIFILTTRFLGKKKKKLFWLASIAPLVSIIL 286
++ ++ H W+ +LG FL+F+ TR L KK KLFW+++IAP+ +++
Sbjct: 718 LEGIFSNRHEIRWE--TTVLGIIFLVFLQFTRHLRLKKPKLFWVSAIAPITCVVV 876
>TC83044 similar to PIR|T48901|T48901 sulfate transporter ATST1 [imported] -
Arabidopsis thaliana, partial (50%)
Length = 1278
Score = 305 bits (782), Expect = 3e-83
Identities = 162/330 (49%), Positives = 223/330 (67%), Gaps = 2/330 (0%)
Frame = +1
Query: 25 PEPPTAWNMVTDSVKKTISQFPRK-LSYLKDQPCNT-LMSFLQGIFPILSWGRNYTAAKF 82
P P + S+K+T FP L K+QP + L+ LQ FPI W +YT
Sbjct: 292 PPPQPFLKSMKYSMKETF--FPDDPLRRFKNQPASKKLVLGLQYFFPIFEWAPSYTFHFL 465
Query: 83 RKDILAGLTIASLCIPQSIGYATLAHLDPQYGLYTSVVPPLIYAVMGTSREVAVGPVAVV 142
+ D++AG+TIASL IPQ I YA LA+L P GLY+S +PPLIYA+MG+SR++AVG VAV
Sbjct: 466 KSDLIAGITIASLAIPQGISYAKLANLPPILGLYSSFIPPLIYAMMGSSRDLAVGTVAVG 645
Query: 143 SLLLFSMVQKLVDPAVDPVAYTKLVFLTTLFAGIFQTAFGLFRLGFLVDFLSHAAIVGFV 202
SLL+ SM+ V+P +P + L F T FAG+ Q + GLFRLGF+VDFLSHAAIVGF+
Sbjct: 646 SLLMGSMLANEVNPTQNPKLFLHLAFTATFFAGLLQASLGLFRLGFIVDFLSHAAIVGFM 825
Query: 203 AGAAIVIGLQQFKGLLGITHFTTKTDIISVMKAVWEALHNPWQPRNFILGSSFLIFILTT 262
GAA V+ LQQ K +LG+ HFT DI+SVM++V+ H W+ + +LG F+ F+L T
Sbjct: 826 GGAATVVCLQQLKSILGLEHFTHAADIVSVMRSVFTQTHQ-WRWESAVLGFCFIFFLLVT 1002
Query: 263 RFLGKKKKKLFWLASIAPLVSIILSTLIVFLTRADKSGVKIVKHVKGGLNPSSLHELDFN 322
R+ KK+ K FW++++ PL S+IL +L+V+ T A+ GV+++ +K GLNP SL +L F
Sbjct: 1003RYFSKKQPKFFWVSAMTPLASVILGSLLVYFTHAEHHGVQVIGELKKGLNPPSLTDLVFV 1182
Query: 323 NPHVGEAAKIGLVVAVVALTESIAVGRSFA 352
+P++ A K GL+V ++AL E IAVGRSFA
Sbjct: 1183SPYMTTAIKTGLIVGIIALAEGIAVGRSFA 1272
>TC91610 weakly similar to PIR|T50022|T50022 sulfate transporter -
Arabidopsis thaliana, partial (21%)
Length = 756
Score = 281 bits (719), Expect = 5e-76
Identities = 140/171 (81%), Positives = 159/171 (92%)
Frame = +3
Query: 465 FFGVLFASVEIGLLAAVMISFLKIILISIRPDTEALGKLPGTSLFCDVYQYPMAVQIPGV 524
FFGVLFASVEIGLL AV+ISF KII+ISIRP TE LGKLPGT LFCDV QYPMA+QIPGV
Sbjct: 3 FFGVLFASVEIGLLVAVVISFAKIIVISIRPSTETLGKLPGTDLFCDVDQYPMAIQIPGV 182
Query: 525 VVIRVKSALLCFANANFVRERIMKWVTQEESKDDKGNSTNTIQLVILETSNLVDIDTSGI 584
++IR+KSALLCFANANFV+ERI+KWVTQ+ +DDKGNS +TIQLVIL+TSNLV+IDTSGI
Sbjct: 183 MIIRMKSALLCFANANFVKERIIKWVTQKGLEDDKGNSKSTIQLVILDTSNLVNIDTSGI 362
Query: 585 ASLEEMQKVLISNGKQLAIANPRWQVIHKLKVSNFVSKIGGRIYLTVEEAI 635
AS+EE+ K L ++GKQLAIANPRWQVIHKLKVSNFVSKIGGR+YLTVEEA+
Sbjct: 363 ASMEELYKCLSTHGKQLAIANPRWQVIHKLKVSNFVSKIGGRVYLTVEEAV 515
>TC84766 similar to SP|P53392|SUT2_STYHA High affinity sulphate transporter
2. {Stylosanthes hamata}, partial (37%)
Length = 873
Score = 240 bits (612), Expect = 1e-63
Identities = 118/203 (58%), Positives = 151/203 (74%)
Frame = +1
Query: 64 LQGIFPILSWGRNYTAAKFRKDILAGLTIASLCIPQSIGYATLAHLDPQYGLYTSVVPPL 123
LQ +FPIL WGR Y F+ D+++GLTIASLCIPQ I YA LA+L+PQY LYTS V PL
Sbjct: 232 LQSVFPILEWGRGYNLKSFKGDLISGLTIASLCIPQDIAYAKLANLEPQYALYTSFVAPL 411
Query: 124 IYAVMGTSREVAVGPVAVVSLLLFSMVQKLVDPAVDPVAYTKLVFLTTLFAGIFQTAFGL 183
+YA MG+SR++A+GPVAVVSLLL S++ + + P Y L F +T FAG+ Q A G+
Sbjct: 412 VYAFMGSSRDIAIGPVAVVSLLLGSLLSEEISDFKSP-EYLALAFTSTFFAGVVQMALGV 588
Query: 184 FRLGFLVDFLSHAAIVGFVAGAAIVIGLQQFKGLLGITHFTTKTDIISVMKAVWEALHNP 243
+LGFL+DFLSHAAIVGF+ GAAI I LQQ KGLLGI FT KTDI+SVM +V++A H+
Sbjct: 589 LKLGFLIDFLSHAAIVGFMGGAAITIALQQLKGLLGIKKFTKKTDIVSVMTSVFKAAHHG 768
Query: 244 WQPRNFILGSSFLIFILTTRFLG 266
W + I+G SFL F+ T+++G
Sbjct: 769 WNWQTIIIGLSFLGFLFITKYIG 837
>AW687663 homologue to GP|13487717|gb sulfate transporter 2 {Lycopersicon
esculentum}, partial (23%)
Length = 526
Score = 199 bits (506), Expect = 3e-51
Identities = 93/146 (63%), Positives = 121/146 (82%)
Frame = +1
Query: 338 VVALTESIAVGRSFASIKGYQLDGNKEMMSIGFSNIIGSLTSCYVATGSFSRTAVNYAAG 397
++ALTE+IA+GR+FAS+K YQLDGNKEM+++G N++GS+TSCYVATGSFSR+AVN+ AG
Sbjct: 1 MIALTEAIAIGRTFASMKDYQLDGNKEMVALGAMNVVGSMTSCYVATGSFSRSAVNFMAG 180
Query: 398 CETLISNIVMAITVLISLQFFTKLLYYTPTAIIASVILSALPGLIDIPEACKIWKVDKLD 457
CET +SNIVM++ V ++LQF T L YTP AI+AS+I+ A+ L+D A IWK+DK D
Sbjct: 181 CETAVSNIVMSVVVFLTLQFITPLFKYTPNAILASIIICAVINLVDYKAAILIWKIDKFD 360
Query: 458 FLACAGAFFGVLFASVEIGLLAAVMI 483
F+AC GAFFGV+FASVEIGLL A +
Sbjct: 361 FVACMGAFFGVVFASVEIGLLIACFL 438
>TC92748 similar to SP|P53393|SUT3_STYHA Low affinity sulphate transporter
3. {Stylosanthes hamata}, partial (20%)
Length = 536
Score = 165 bits (418), Expect(2) = 4e-48
Identities = 83/141 (58%), Positives = 111/141 (77%), Gaps = 7/141 (4%)
Frame = +2
Query: 228 DIISVMKAVWEALHNP------WQPRNFILGSSFLIFILTTRFLGKKKKKLFWLASIAPL 281
D +SV+ +V+++LH W P NF+LG SFLIF+L TRF+ +KKKKLFWL +IAPL
Sbjct: 2 DAVSVLVSVYKSLHQQITSEEKWSPLNFVLGCSFLIFLLVTRFIARKKKKLFWLPAIAPL 181
Query: 282 VSIILSTLIVFLTRADKSGVKIVKHVKGGLNPSSLHELDFNNPHVGEAAKIGLVVAVVAL 341
+S+ILSTLIV+L++ADK G+ I+KHVKGGLN SS+H+L F+ +VG+AAKIGLV AV+AL
Sbjct: 182 LSVILSTLIVYLSKADKQGINIIKHVKGGLNQSSVHQLQFHGQNVGQAAKIGLVCAVIAL 361
Query: 342 TESIAVGRSFASIK-GYQLDG 361
TE++A G SFA ++ LDG
Sbjct: 362 TEAMAGGPSFAFLQXDTHLDG 424
Score = 45.1 bits (105), Expect(2) = 4e-48
Identities = 22/40 (55%), Positives = 27/40 (67%)
Frame = +3
Query: 359 LDGNKEMMSIGFSNIIGSLTSCYVATGSFSRTAVNYAAGC 398
L GN+EM+++G N G+LTS YV GS SRTAV GC
Sbjct: 417 LMGNREMVAMGIMNXAGTLTSWYVXPGSXSRTAVISMXGC 536
>TC83848 similar to SP|P53393|SUT3_STYHA Low affinity sulphate transporter
3. {Stylosanthes hamata}, partial (21%)
Length = 769
Score = 169 bits (428), Expect(2) = 2e-45
Identities = 88/163 (53%), Positives = 116/163 (70%), Gaps = 3/163 (1%)
Frame = +3
Query: 37 SVKKTISQFPRKLSYLKDQPCNTLM--SFLQGIFPILSWGRNYTAAKFRKDILAGLTIAS 94
S+K+T+ KL + + SFLQ +FPIL W ++YT +KF+ D+LAGLT+AS
Sbjct: 246 SLKETLLPHGNKLCFSSKNKSFLALAYSFLQSLFPILVWLKDYTISKFKDDLLAGLTLAS 425
Query: 95 LCIPQSIGYATLAHLDPQYGLYTSVVPPLIYAVMGTSREVAVGPVAVVSLLLFSMVQKLV 154
LCIPQSIGYA+LA +DPQYGLYTS+VPPLIYAVMG+SR++A+GPVAVVS+LL S+V ++
Sbjct: 426 LCIPQSIGYASLAKVDPQYGLYTSIVPPLIYAVMGSSRDIAIGPVAVVSMLLSSLVTNVI 605
Query: 155 DPAVDPVAYTKLVFLTTLFAGIFQTAFGLFRLGF-LVDFLSHA 196
DP +P AY +F L F + F++GF L FLSHA
Sbjct: 606 DPVANPHAYRDFIFTVHLLHRNFSSCIWYFQVGFSLWIFLSHA 734
Score = 41.2 bits (95), Expect = 0.001
Identities = 18/35 (51%), Positives = 23/35 (65%)
Frame = +1
Query: 170 TTLFAGIFQTAFGLFRLGFLVDFLSHAAIVGFVAG 204
+T F GIFQ AFG+FRLGFL F H + ++ G
Sbjct: 652 STFFTGIFQAAFGIFRLGFLCGFFFHMPALVWIHG 756
Score = 32.0 bits (71), Expect(2) = 2e-45
Identities = 13/26 (50%), Positives = 19/26 (73%), Gaps = 1/26 (3%)
Frame = +2
Query: 7 LDIEKNTQEV-RSQWVLNAPEPPTAW 31
L IE +T ++ RS+WVL++P PP W
Sbjct: 152 LHIEDSTSQIERSKWVLDSPNPPPLW 229
>TC93814 similar to PIR|B86365|B86365 probable sulphate transporter protein
[imported] - Arabidopsis thaliana, partial (30%)
Length = 671
Score = 179 bits (455), Expect = 2e-45
Identities = 95/200 (47%), Positives = 131/200 (65%), Gaps = 20/200 (10%)
Frame = +2
Query: 311 LNPSSLHELDFNNPHVGEAAK-----------IGLVVA----VVAL-----TESIAVGRS 350
+NPSS + L F+ H+G K +G + V+ L E IAVGR+
Sbjct: 2 INPSSWNMLKFHGSHLGLVVKTWHYHWHFVPHLGFINGKNG*VICLFKKNKQEGIAVGRT 181
Query: 351 FASIKGYQLDGNKEMMSIGFSNIIGSLTSCYVATGSFSRTAVNYAAGCETLISNIVMAIT 410
FA++ Y++DGNKEMM+IG N++GS TSCYV TG+FSR+AVN AG +T SNIVM++T
Sbjct: 182 FAALGNYKVDGNKEMMAIGCMNVVGSFTSCYVTTGAFSRSAVNNNAGAKTAASNIVMSVT 361
Query: 411 VLISLQFFTKLLYYTPTAIIASVILSALPGLIDIPEACKIWKVDKLDFLACAGAFFGVLF 470
V+++L F L YTP ++ ++I++A+ GLIDIP AC IWK+DK DFL AFFGV+F
Sbjct: 362 VMVTLLFLMPLFQYTPNVVLGAIIVTAVIGLIDIPAACHIWKIDKFDFLVMLTAFFGVVF 541
Query: 471 ASVEIGLLAAVMISFLKIIL 490
SV++GL AV +S I+L
Sbjct: 542 ISVQLGLAVAVGLSTFXILL 601
>BI267593 similar to PIR|B86365|B86 probable sulphate transporter protein
[imported] - Arabidopsis thaliana, partial (32%)
Length = 646
Score = 163 bits (413), Expect(2) = 2e-42
Identities = 76/138 (55%), Positives = 107/138 (77%)
Frame = +3
Query: 68 FPILSWGRNYTAAKFRKDILAGLTIASLCIPQSIGYATLAHLDPQYGLYTSVVPPLIYAV 127
FPIL WG Y+ F+ D+++GLTIASL IPQ I YA LA+L P GLY+S VPPLIYAV
Sbjct: 168 FPILQWGSIYSFKLFKSDLVSGLTIASLAIPQGISYAKLANLPPIVGLYSSFVPPLIYAV 347
Query: 128 MGTSREVAVGPVAVVSLLLFSMVQKLVDPAVDPVAYTKLVFLTTLFAGIFQTAFGLFRLG 187
+G+SR++AVGPV++ SL+L SM+++ V P+ +PV + +L +T FAG+FQ + G+ RLG
Sbjct: 348 LGSSRDLAVGPVSIASLVLGSMLRQEVSPSAEPVLFLQLALTSTFFAGLFQASLGILRLG 527
Query: 188 FLVDFLSHAAIVGFVAGA 205
F++DFLS A ++GF+AG+
Sbjct: 528 FIIDFLSKAILIGFMAGS 581
Score = 28.1 bits (61), Expect(2) = 2e-42
Identities = 14/23 (60%), Positives = 15/23 (64%)
Frame = +1
Query: 204 GAAIVIGLQQFKGLLGITHFTTK 226
GAAI+ Q K LLGITH T K
Sbjct: 577 GAAIIGVFAQLKSLLGITHXTHK 645
>AL378429 similar to GP|9294446|dbj sulfate transporter {Arabidopsis
thaliana}, partial (22%)
Length = 473
Score = 148 bits (374), Expect = 6e-36
Identities = 72/150 (48%), Positives = 104/150 (69%)
Frame = +3
Query: 185 RLGFLVDFLSHAAIVGFVAGAAIVIGLQQFKGLLGITHFTTKTDIISVMKAVWEALHNPW 244
RLGF++DFLS A +VGF+AGAAI++ LQQ KGLLGI HFT K II V+ +V++ + W
Sbjct: 27 RLGFVIDFLSKATLVGFMAGAAIIVSLQQLKGLLGIVHFTNKMQIIPVLVSVYKQ-KDEW 203
Query: 245 QPRNFILGSSFLIFILTTRFLGKKKKKLFWLASIAPLVSIILSTLIVFLTRADKSGVKIV 304
+ ++G FL F+LTTR + +K KLFW ++ APL S+ILST++VFL R + ++
Sbjct: 204 SWQTIVMGFGFLAFLLTTRHISLRKPKLFWASAAAPLTSVILSTILVFLLRNKAHHISVI 383
Query: 305 KHVKGGLNPSSLHELDFNNPHVGEAAKIGL 334
H+ G+NP S++ L FN P++ A K G+
Sbjct: 384 GHLAKGVNPPSVNLLYFNGPYLALAIKTGI 473
>AW685206 similar to GP|9955547|emb sulfate transporter {Arabidopsis
thaliana}, partial (25%)
Length = 540
Score = 132 bits (333), Expect = 3e-31
Identities = 69/178 (38%), Positives = 107/178 (59%)
Frame = +3
Query: 349 RSFASIKGYQLDGNKEMMSIGFSNIIGSLTSCYVATGSFSRTAVNYAAGCETLISNIVMA 408
++ A+ GY+LD N+E++ +G SN++GS S Y TGSFSR+AVN+ +G ++ +S IV
Sbjct: 3 KALAAKNGYELDSNQELVGLGVSNVLGSFFSAYPTTGSFSRSAVNHESGAKSGVSAIVSG 182
Query: 409 ITVLISLQFFTKLLYYTPTAIIASVILSALPGLIDIPEACKIWKVDKLDFLACAGAFFGV 468
I + +L F T L P + +A++++SA+ GL+D EA +W+VDK DFL
Sbjct: 183 IIITCALLFLTPLFENIPQSALAAIVISAVIGLVDYDEAIFLWRVDKKDFLLWILTSTTT 362
Query: 469 LFASVEIGLLAAVMISFLKIILISIRPDTEALGKLPGTSLFCDVYQYPMAVQIPGVVV 526
LF +EIG++ V S +I S P LG+LPGT++ + QY A G+V+
Sbjct: 363 LFLGIEIGVMVGVGASLAFVIHESANPHIAVLGRLPGTTVXRNXKQYXEAYTYNGIVI 536
>TC83238 weakly similar to PIR|T49069|T49069 sulfate transporter (ATST1) -
Arabidopsis thaliana, partial (21%)
Length = 778
Score = 124 bits (311), Expect = 1e-28
Identities = 64/163 (39%), Positives = 106/163 (64%), Gaps = 1/163 (0%)
Frame = +2
Query: 475 IGLLAAVMISFLKIILISIRPDTEALGKLPGTSLFCDVYQYPMAVQIPGVVVIRVKSALL 534
IGL+ AV IS L+++L RP T LG +P + + +V Y A +PG++++ + + +
Sbjct: 2 IGLIIAVAISVLRVLLFVARPRTFVLGNIPNSGAYRNVEHYQNAHHVPGILILEIDAPIF 181
Query: 535 CFANANFVRERIMKWVTQEESKDDKGNSTNTIQLVILETSNLVDIDTSGIASLEEMQKVL 594
FANA+++RERI +W+ +EE + + T+ +Q VI++ + + +IDTSGI+ LEE +K +
Sbjct: 182 -FANASYLRERITRWINEEEERIEGAGKTS-LQYVIMDMTAVANIDTSGISMLEEFKKTV 355
Query: 595 ISNGKQLAIANPRWQVIHKLKVSNFVSKIGGR-IYLTVEEAIG 636
G QL + NP +V KL S F+ +IG + +Y+TVEEA+G
Sbjct: 356 DRKGLQLVMVNPGSEVTKKLNKSKFLDEIGHKWVYVTVEEAVG 484
>TC90755 similar to SP|P53391|SUT1_STYHA High affinity sulphate transporter
1. {Stylosanthes hamata}, partial (24%)
Length = 847
Score = 117 bits (292), Expect = 2e-26
Identities = 67/152 (44%), Positives = 99/152 (65%), Gaps = 1/152 (0%)
Frame = +1
Query: 485 FLKIILISIRPDTEALGKLPGTSLFCDVYQYPMAVQIPGVVVIRVKSALLCFANANFVRE 544
F + +L RP T LGKLPGT+++ ++ QYP A QIPG++++RV SA+ F+N+N++++
Sbjct: 22 FCQNLLQVTRPKTAVLGKLPGTTVYRNILQYPKAAQIPGMLIVRVDSAIY-FSNSNYIKD 198
Query: 545 RIMKWVTQEESKDDKGNSTNTIQLVILETSNLVDIDTSGIASLEEMQKVLISNGKQLAIA 604
RI+KW+T EE + +IQ +I+E S + DIDTSGI S E++ K L QL +A
Sbjct: 199 RILKWLTDEEIL-RTSSEYPSIQHLIVEMSPVTDIDTSGIHSFEDLLKSLKKRDIQLLLA 375
Query: 605 NPRWQVIHKLKVSNFVSKIG-GRIYLTVEEAI 635
NP VI KL S IG +I+LTV +A+
Sbjct: 376 NPGPIVIEKLHASKLSDLIGEDKIFLTVGDAV 471
>AW560689 similar to GP|9294446|dbj| sulfate transporter {Arabidopsis
thaliana}, partial (13%)
Length = 276
Score = 63.5 bits (153), Expect(2) = 2e-16
Identities = 31/46 (67%), Positives = 37/46 (80%)
Frame = +3
Query: 89 GLTIASLCIPQSIGYATLAHLDPQYGLYTSVVPPLIYAVMGTSREV 134
GLTIASL IPQ I YA LA+L P GLY+S+VPP IYA++G+SR V
Sbjct: 3 GLTIASLAIPQGISYAKLANLPPIIGLYSSIVPP*IYALLGSSRHV 140
Score = 40.4 bits (93), Expect(2) = 2e-16
Identities = 19/44 (43%), Positives = 28/44 (63%)
Frame = +1
Query: 136 VGPVAVVSLLLFSMVQKLVDPAVDPVAYTKLVFLTTLFAGIFQT 179
VGPV++ SL++ SM+ + V +P Y +L F T AG+FQT
Sbjct: 145 VGPVSIASLVMGSMLSETVFFTQNPTLYLQLAFTATFVAGLFQT 276
>TC81621 similar to PIR|T49069|T49069 sulfate transporter (ATST1) -
Arabidopsis thaliana, partial (14%)
Length = 863
Score = 61.6 bits (148), Expect = 9e-10
Identities = 38/81 (46%), Positives = 48/81 (58%), Gaps = 2/81 (2%)
Frame = +2
Query: 37 SVKKTISQFPRK-LSYLKDQPCNT-LMSFLQGIFPILSWGRNYTAAKFRKDILAGLTIAS 94
SVK+T+ FP L K+QP + + LQ PIL W YT + D++AG+TIAS
Sbjct: 515 SVKETL--FPDDPLRKFKNQPASRRFVLALQHFLPILEWAPQYTLQFLKSDLIAGITIAS 688
Query: 95 LCIPQSIGYATLAHLDPQYGL 115
L IPQ I YA LA+L P GL
Sbjct: 689 LAIPQGISYAKLANLPPILGL 751
>BG646860 similar to GP|13487717|gb| sulfate transporter 2 {Lycopersicon
esculentum}, partial (13%)
Length = 752
Score = 46.2 bits (108), Expect(2) = 5e-08
Identities = 22/31 (70%), Positives = 25/31 (79%)
Frame = +3
Query: 71 LSWGRNYTAAKFRKDILAGLTIASLCIPQSI 101
L+ GR+Y KFR DI+AGLTIASLCIPQ I
Sbjct: 486 LTGGRSYNLKKFRGDIIAGLTIASLCIPQVI 578
Score = 29.3 bits (64), Expect(2) = 5e-08
Identities = 12/15 (80%), Positives = 13/15 (86%)
Frame = +2
Query: 101 IGYATLAHLDPQYGL 115
IGY+ LAHL PQYGL
Sbjct: 668 IGYSKLAHLAPQYGL 712
>AL378430 similar to GP|9294446|dbj| sulfate transporter {Arabidopsis
thaliana}, partial (13%)
Length = 475
Score = 51.2 bits (121), Expect = 1e-06
Identities = 27/72 (37%), Positives = 45/72 (62%), Gaps = 1/72 (1%)
Frame = +2
Query: 565 TIQLVILETSNLVDIDTSGIASLEEMQKVLISNGKQLAIANPRWQVIHKLKVSNFVSKIG 624
++Q +IL+ + + IDTSG+ +L E++K+L QL + NP V+ KL +S + G
Sbjct: 23 SLQCIILDMTAVTAIDTSGLETLGELRKMLEKRSLQLVLVNPVGNVMEKLHMSKILDTFG 202
Query: 625 GR-IYLTVEEAI 635
+ +YLTV EA+
Sbjct: 203 FKGLYLTVGEAV 238
>BF635989 weakly similar to GP|9294446|dbj| sulfate transporter {Arabidopsis
thaliana}, partial (9%)
Length = 655
Score = 39.3 bits (90), Expect = 0.005
Identities = 23/56 (41%), Positives = 34/56 (60%), Gaps = 1/56 (1%)
Frame = +2
Query: 581 TSGIASLEEMQKVLISNGKQLAIANPRWQVIHKLKVSNFVSKIGGR-IYLTVEEAI 635
TSGI +L E+++ L QL +ANP V+ KL SN + G + +YL+V EA+
Sbjct: 2 TSGIDTLCELRRRLEQKSLQLVLANPVGNVMEKLHESNILDSFGMKGLYLSVGEAV 169
>TC79406 weakly similar to PIR|T04879|T04879 hypothetical protein F18F4.50 -
Arabidopsis thaliana, partial (26%)
Length = 1053
Score = 32.0 bits (71), Expect = 0.75
Identities = 24/84 (28%), Positives = 38/84 (44%)
Frame = +2
Query: 401 LISNIVMAITVLISLQFFTKLLYYTPTAIIASVILSALPGLIDIPEACKIWKVDKLDFLA 460
L+ +V +LI T + YT +I S +S + +P K+WK + FL
Sbjct: 395 LLFKLVYFTFLLIFSLLSTSAIVYTTASIFTSKDVSFKKVMKIVP---KVWKRLMITFLC 565
Query: 461 CAGAFFGVLFASVEIGLLAAVMIS 484
AFF F + + +L A+ IS
Sbjct: 566 AYAAFFAYNFITFFVIILLAITIS 637
>TC87340 similar to GP|15215816|gb|AAK91453.1 At2g26990/T20P8.4 {Arabidopsis
thaliana}, partial (54%)
Length = 842
Score = 30.8 bits (68), Expect = 1.7
Identities = 24/80 (30%), Positives = 38/80 (47%), Gaps = 5/80 (6%)
Frame = -3
Query: 253 SSFLIFILTTRFLGKKKKKLFWLASIAPLVSIIL-----STLIVFLTRADKSGVKIVKHV 307
S F + + +LGK +K FW++ L +IL L++FL R KS IV H+
Sbjct: 306 SPFCLVLFHAHYLGKTRKCTFWISFNQTL*IVILILNVNILLLLFLIRVLKS---IVLHI 136
Query: 308 KGGLNPSSLHELDFNNPHVG 327
G + + ++ NP G
Sbjct: 135 SVG-SHDCVKSIELENPS*G 79
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.139 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,284,241
Number of Sequences: 36976
Number of extensions: 232934
Number of successful extensions: 1533
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 1509
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1521
length of query: 641
length of database: 9,014,727
effective HSP length: 102
effective length of query: 539
effective length of database: 5,243,175
effective search space: 2826071325
effective search space used: 2826071325
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0306.8


