

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0306.5
(165 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
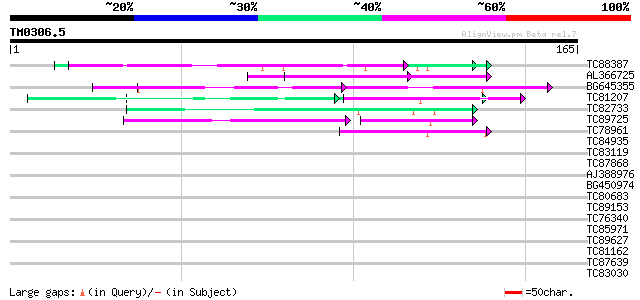
Score E
Sequences producing significant alignments: (bits) Value
TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 63 5e-11
AL366725 60 5e-10
BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.... 60 5e-10
TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 52 1e-07
TC82733 similar to GP|10177404|dbj|BAB10535. gene_id:K24M7.12~pi... 50 4e-07
TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.... 47 5e-06
TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Ar... 45 2e-05
TC84935 similar to PIR|G96631|G96631 probable RNA-binding protei... 38 0.002
TC83119 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 33 0.040
TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 33 0.052
AJ388976 similar to PIR|E84638|E84 probable RSZp22 splicing fact... 32 0.15
BG450974 similar to PIR|T05112|T05 splicing factor 9G8-like SR p... 31 0.20
TC80683 homologue to GP|8777424|dbj|BAA97014.1 gb|AAF56406.1~gen... 30 0.34
TC89153 similar to GP|18855061|gb|AAL79753.1 putative RNA helica... 30 0.44
TC76340 homologue to SP|Q9SM09|VATA_CITUN Vacuolar ATP synthase ... 30 0.58
TC85971 similar to GP|19424073|gb|AAL87350.1 unknown protein {Ar... 29 0.98
TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pec... 29 0.98
TC81162 weakly similar to PIR|T47673|T47673 hypothetical protein... 29 0.98
TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.... 28 1.3
TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA... 28 1.3
>TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (96%)
Length = 1286
Score = 63.2 bits (152), Expect = 5e-11
Identities = 36/133 (27%), Positives = 53/133 (39%), Gaps = 6/133 (4%)
Frame = +1
Query: 14 KDVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTG 73
+D C C + G + C + C C+ GH+A C T
Sbjct: 412 QDNLCKNCKRPGHYVRECPNVAV-CHNCSLPGHIASECSTKS------------------ 534
Query: 74 NKTSGKIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKCKIKG------RLCFNCN 127
C+ CK+ GH + CP+ G +C CG+ GH A +C + RLC NC
Sbjct: 535 --------LCWNCKEPGHMASSCPNEG-ICHTCGKAGHRARECTVPQKPPGDLRLCNNCY 687
Query: 128 QPGHFSRDCKAPK 140
+ GH + +C K
Sbjct: 688 KQGHIAVECTNEK 726
Score = 62.4 bits (150), Expect = 8e-11
Identities = 40/141 (28%), Positives = 54/141 (37%), Gaps = 22/141 (15%)
Frame = +1
Query: 18 CFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTS 77
C+ C + G A++C G C C + GH A C Q P D ++ N G
Sbjct: 538 CWNCKEPGHMASSCPNEGI-CHTCGKAGHRARECTVPQKPPGDLRLCNNCYKQGHIAVEC 714
Query: 78 GKIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKCK-------------------- 117
C C+K GH CP+ P+C C GH A +C
Sbjct: 715 TNEKACNNCRKTGHLARDCPND-PICNLCNISGHVARQCPKSNVIGDRGGGGSLRGGYRD 891
Query: 118 --IKGRLCFNCNQPGHFSRDC 136
+ +C +C Q GH SRDC
Sbjct: 892 GGFRDVVCRSCQQFGHMSRDC 954
Score = 53.9 bits (128), Expect = 3e-08
Identities = 38/105 (36%), Positives = 49/105 (46%), Gaps = 6/105 (5%)
Frame = +1
Query: 18 CFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGT---GN 74
C C K G A +C P C CN GH+A C S + G+ GG+ G
Sbjct: 730 CNNCRKTGHLARDCPN-DPICNLCNISGHVARQCPK-------SNVIGDRGGGGSLRGGY 885
Query: 75 KTSG-KIVTCYRCKKVGHYTNKCPDGGPL--CFNCGEIGHYASKC 116
+ G + V C C++ GH + C GGPL C NCG GH A +C
Sbjct: 886 RDGGFRDVVCRSCQQFGHMSRDCM-GGPLMICQNCGGRGHQAYEC 1017
Score = 32.0 bits (71), Expect = 0.12
Identities = 16/42 (38%), Positives = 21/42 (49%), Gaps = 2/42 (4%)
Frame = +1
Query: 12 GDKDVTCFCCGKFGQFANNCSVIGPR--CFKCNREGHLAVNC 51
G +DV C C +FG + +C + GP C C GH A C
Sbjct: 895 GFRDVVCRSCQQFGHMSRDC-MGGPLMICQNCGGRGHQAYEC 1017
>AL366725
Length = 485
Score = 59.7 bits (143), Expect = 5e-10
Identities = 23/60 (38%), Positives = 31/60 (51%)
Frame = +2
Query: 81 VTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKCKIKGRLCFNCNQPGHFSRDCKAPK 140
+ C+ + GH +N CP C C + GH + CK +CFNCN+ GH CK PK
Sbjct: 302 IVCFNYGEKGHKSNVCPKEIKKCVRCDKKGHIVADCKRNDIVCFNCNEEGHIGSQCKQPK 481
Score = 45.4 bits (106), Expect = 1e-05
Identities = 20/48 (41%), Positives = 23/48 (47%)
Frame = +2
Query: 70 GGTGNKTSGKIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKCK 117
G N +I C RC K GH C +CFNC E GH S+CK
Sbjct: 329 GHKSNVCPKEIKKCVRCDKKGHIVADCKRNDIVCFNCNEEGHIGSQCK 472
Score = 38.9 bits (89), Expect = 0.001
Identities = 19/82 (23%), Positives = 35/82 (42%)
Frame = +2
Query: 15 DVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGN 74
++ CF G+ G +N C +C +C+++GH+ +CK ND
Sbjct: 299 EIVCFNYGEKGHKSNVCPKEIKKCVRCDKKGHIVADCKR------------ND------- 421
Query: 75 KTSGKIVTCYRCKKVGHYTNKC 96
+ C+ C + GH ++C
Sbjct: 422 ------IVCFNCNEEGHIGSQC 469
Score = 35.0 bits (79), Expect = 0.014
Identities = 14/35 (40%), Positives = 16/35 (45%)
Frame = +2
Query: 18 CFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCK 52
C C K G +C CF CN EGH+ CK
Sbjct: 368 CVRCDKKGHIVADCKRNDIVCFNCNEEGHIGSQCK 472
>BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.5 [imported]
- Arabidopsis thaliana, partial (5%)
Length = 627
Score = 59.7 bits (143), Expect = 5e-10
Identities = 34/125 (27%), Positives = 53/125 (42%), Gaps = 4/125 (3%)
Frame = -1
Query: 38 CFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCP 97
C C H + C L+ P + T SGK CY+C++ GH+ + CP
Sbjct: 627 CSNCGGSDHSSAQCLHLRNPPEQTAGGAYVNTVSGSGGASGK---CYKCQQPGHWASNCP 457
Query: 98 DGGPLCFNCGEIGHYASKCKIKGRLCFNCNQPGHFSRDC----KAPKGESSGNTGKGKQL 153
G G + C + CNQPGH++ +C AP+ + NTG+G+
Sbjct: 456 SMSAANRVSGGSGGASGNC-------YKCNQPGHWANNCPNMSAAPQSHGNSNTGQGRYG 298
Query: 154 AAEER 158
A+ +
Sbjct: 297 IAQNQ 283
Score = 49.3 bits (116), Expect = 7e-07
Identities = 24/77 (31%), Positives = 37/77 (47%), Gaps = 3/77 (3%)
Frame = -1
Query: 25 GQFANNCSVIGP---RCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIV 81
G + N S G +C+KC + GH A NC ++ N +GG+G +
Sbjct: 549 GAYVNTVSGSGGASGKCYKCQQPGHWASNCPSMSAA--------NRVSGGSGGASGN--- 403
Query: 82 TCYRCKKVGHYTNKCPD 98
CY+C + GH+ N CP+
Sbjct: 402 -CYKCNQPGHWANNCPN 355
Score = 38.1 bits (87), Expect = 0.002
Identities = 20/66 (30%), Positives = 29/66 (43%), Gaps = 13/66 (19%)
Frame = -1
Query: 18 CFCCGKFGQFANNCSVIGP-------------RCFKCNREGHLAVNCKTLQGGPSDSKMK 64
C+ C + G +A+NC + C+KCN+ GH A NC + P +
Sbjct: 501 CYKCQQPGHWASNCPSMSAANRVSGGSGGASGNCYKCNQPGHWANNCPNMSAAP---QSH 331
Query: 65 GNDATG 70
GN TG
Sbjct: 330 GNSNTG 313
>TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (39%)
Length = 630
Score = 51.6 bits (122), Expect = 1e-07
Identities = 32/105 (30%), Positives = 41/105 (38%)
Frame = +3
Query: 35 GPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTN 94
G C+ C GH+A +C + ND +GG G + CY C H+
Sbjct: 342 GGGCYTCGDTGHIARDCDR------SDRNDRNDRSGGGGGGDRDR--ACYTCGSFEHFAR 497
Query: 95 KCPDGGPLCFNCGEIGHYASKCKIKGRLCFNCNQPGHFSRDCKAP 139
C GG N G G Y G C+ C GH +RDC P
Sbjct: 498 DCMRGGGNNNNGG--GGYGGG----GTSCYRCGGVGHIARDCATP 614
Score = 43.1 bits (100), Expect = 5e-05
Identities = 28/91 (30%), Positives = 36/91 (38%)
Frame = +3
Query: 6 TGG*PIGDKDVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKG 65
+GG GD+D C+ CG F FA +C ++GG G
Sbjct: 426 SGGGGGGDRDRACYTCGSFEHFARDC----------------------MRGG-------G 518
Query: 66 NDATGGTGNKTSGKIVTCYRCKKVGHYTNKC 96
N+ GG G G +CYRC VGH C
Sbjct: 519 NNNNGGGGYGGGG--TSCYRCGGVGHIARDC 605
Score = 39.7 bits (91), Expect = 6e-04
Identities = 21/70 (30%), Positives = 29/70 (41%), Gaps = 17/70 (24%)
Frame = +3
Query: 98 DGGPLCFNCGEIGHYASKCKI-----------------KGRLCFNCNQPGHFSRDCKAPK 140
+GG C+ CG+ GH A C + R C+ C HF+RDC +
Sbjct: 336 NGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCM--R 509
Query: 141 GESSGNTGKG 150
G + N G G
Sbjct: 510 GGGNNNNGGG 539
>TC82733 similar to GP|10177404|dbj|BAB10535.
gene_id:K24M7.12~pir||S42136~similar to unknown protein
{Arabidopsis thaliana}, partial (57%)
Length = 710
Score = 50.1 bits (118), Expect = 4e-07
Identities = 33/114 (28%), Positives = 44/114 (37%), Gaps = 12/114 (10%)
Frame = +3
Query: 35 GPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTN 94
G CF C H+A C T K C RC++ GH
Sbjct: 273 GDSCFICKGLDHIAKFC--------------------TQKAEWEKNKICLRCRRRGHRAQ 392
Query: 95 KCPDGG-----PLCFNCGEIGHYASKC----KIKGRL---CFNCNQPGHFSRDC 136
CPDGG +NCG+ GH + C + G + CF C + GH S++C
Sbjct: 393 NCPDGGSKEDFKY*YNCGDNGHSLANCPHPLQEGGTMFAQCFVCKEQGHLSKNC 554
Score = 37.4 bits (85), Expect = 0.003
Identities = 29/111 (26%), Positives = 36/111 (32%), Gaps = 20/111 (18%)
Frame = +3
Query: 17 TCFCCGKFGQFANNCSVIGPR-----CFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGG 71
+CF C A C+ C +C R GH A NC G D K N G
Sbjct: 279 SCFICKGLDHIAKFCTQKAEWEKNKICLRCRRRGHRAQNCPD-GGSKEDFKY*YNCGDNG 455
Query: 72 TG--------NKTSGKIVTCYRCKKVGHYTNKCPDG-------GPLCFNCG 107
+ C+ CK+ GH + CP G C CG
Sbjct: 456 HSLANCPHPLQEGGTMFAQCFVCKEQGHLSKNCPKNAHGIYPKGGCCKICG 608
>TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.150 -
Arabidopsis thaliana, partial (17%)
Length = 378
Score = 46.6 bits (109), Expect = 5e-06
Identities = 22/66 (33%), Positives = 29/66 (43%)
Frame = +1
Query: 34 IGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYT 93
+G C+ C GH+A C + GG G GG G G +CY C + GH+
Sbjct: 4 VGGGCYNCGESGHMARECTSGGGGGG-----GRYGGGGGGGGGGGGGGSCYSCGESGHFA 168
Query: 94 NKCPDG 99
CP G
Sbjct: 169RDCPTG 186
Score = 40.0 bits (92), Expect = 4e-04
Identities = 18/54 (33%), Positives = 24/54 (44%), Gaps = 20/54 (37%)
Frame = +1
Query: 103 CFNCGEIGHYASKCKIKGR--------------------LCFNCNQPGHFSRDC 136
C+NCGE GH A +C G C++C + GHF+RDC
Sbjct: 16 CYNCGESGHMARECTSGGGGGGGRYGGGGGGGGGGGGGGSCYSCGESGHFARDC 177
Score = 32.7 bits (73), Expect = 0.068
Identities = 11/28 (39%), Positives = 16/28 (56%)
Frame = +1
Query: 123 CFNCNQPGHFSRDCKAPKGESSGNTGKG 150
C+NC + GH +R+C + G G G G
Sbjct: 16 CYNCGESGHMARECTSGGGGGGGRYGGG 99
>TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Arabidopsis
thaliana}, partial (71%)
Length = 974
Score = 44.7 bits (104), Expect = 2e-05
Identities = 20/47 (42%), Positives = 26/47 (54%), Gaps = 3/47 (6%)
Frame = +2
Query: 97 PDGGPLCFNCGEIGHYASKCKIKG--RLCFNCNQPGHFSRDCK-APK 140
P G CFNCG GH+A CK C+ C + GH ++CK +PK
Sbjct: 341 PPGSGRCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCKNSPK 481
Score = 35.4 bits (80), Expect = 0.010
Identities = 14/37 (37%), Positives = 20/37 (53%), Gaps = 2/37 (5%)
Frame = +2
Query: 18 CFCCGKFGQFANNCSVIG--PRCFKCNREGHLAVNCK 52
CF CG G +A +C +C++C GH+ NCK
Sbjct: 359 CFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCK 469
Score = 34.7 bits (78), Expect = 0.018
Identities = 18/58 (31%), Positives = 23/58 (39%), Gaps = 2/58 (3%)
Frame = +2
Query: 83 CYRCKKVGHYTNKCPDGG--PLCFNCGEIGHYASKCKIKGRLCFNCNQPGHFSRDCKA 138
C+ C GH+ C G C+ CGE GH CK N P SR ++
Sbjct: 359 CFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCK---------NSPKKLSRHARS 505
Score = 31.6 bits (70), Expect = 0.15
Identities = 19/60 (31%), Positives = 22/60 (36%)
Frame = +2
Query: 37 RCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKC 96
RCF C +GH A +CK G NK CYRC + GH C
Sbjct: 356 RCFNCGIDGHWARDCK----------------AGDWKNK-------CYRCGERGHIEKNC 466
>TC84935 similar to PIR|G96631|G96631 probable RNA-binding protein F8A5.17
[imported] - Arabidopsis thaliana, partial (41%)
Length = 552
Score = 38.1 bits (87), Expect = 0.002
Identities = 17/47 (36%), Positives = 25/47 (53%)
Frame = +2
Query: 104 FNCGEIGHYASKCKIKGRLCFNCNQPGHFSRDCKAPKGESSGNTGKG 150
F+ G G Y + ++ CF C +PGH++RDC P G G+G
Sbjct: 416 FSSGGRGSYGAGDRVGQDDCFKCGRPGHWARDC--PLAGGDGGRGRG 550
Score = 32.0 bits (71), Expect = 0.12
Identities = 18/65 (27%), Positives = 27/65 (40%)
Frame = +2
Query: 56 GGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASK 115
G +D + +G ++GG G+ +G V G CF CG GH+A
Sbjct: 383 GDDADQRYRGGFSSGGRGSYGAGDRV-----------------GQDDCFKCGRPGHWARD 511
Query: 116 CKIKG 120
C + G
Sbjct: 512 CPLAG 526
Score = 28.1 bits (61), Expect = 1.7
Identities = 16/39 (41%), Positives = 18/39 (46%)
Frame = +2
Query: 19 FCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGG 57
F G G + V CFKC R GH A +C L GG
Sbjct: 416 FSSGGRGSYGAGDRVGQDDCFKCGRPGHWARDC-PLAGG 529
>TC83119 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (16%)
Length = 421
Score = 33.5 bits (75), Expect = 0.040
Identities = 12/25 (48%), Positives = 16/25 (64%)
Frame = +3
Query: 83 CYRCKKVGHYTNKCPDGGPLCFNCG 107
C CK+ GHY +CP+ +C NCG
Sbjct: 348 CKNCKRPGHYARECPNVA-VCHNCG 419
Score = 30.8 bits (68), Expect = 0.26
Identities = 9/18 (50%), Positives = 14/18 (77%)
Frame = +3
Query: 119 KGRLCFNCNQPGHFSRDC 136
+ LC NC +PGH++R+C
Sbjct: 336 RDNLCKNCKRPGHYAREC 389
Score = 28.1 bits (61), Expect = 1.7
Identities = 12/25 (48%), Positives = 14/25 (56%)
Frame = +3
Query: 102 LCFNCGEIGHYASKCKIKGRLCFNC 126
LC NC GHYA +C +C NC
Sbjct: 345 LCKNCKRPGHYARECP-NVAVCHNC 416
>TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (91%)
Length = 860
Score = 33.1 bits (74), Expect = 0.052
Identities = 10/23 (43%), Positives = 16/23 (69%)
Frame = +3
Query: 123 CFNCNQPGHFSRDCKAPKGESSG 145
C+ C +PGHF+R+C+ G +G
Sbjct: 336 CYECGEPGHFARECRNRGGGGAG 404
Score = 30.0 bits (66), Expect = 0.44
Identities = 9/18 (50%), Positives = 14/18 (77%)
Frame = +3
Query: 103 CFNCGEIGHYASKCKIKG 120
C+ CGE GH+A +C+ +G
Sbjct: 336 CYECGEPGHFARECRNRG 389
Score = 29.6 bits (65), Expect = 0.58
Identities = 15/54 (27%), Positives = 25/54 (45%)
Frame = +3
Query: 47 LAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCPDGG 100
L+ N ++ GG +G GG G+ + CY C + GH+ +C + G
Sbjct: 243 LSHNSRSGGGGGGGGGGRGRGGGGGGGSD-----LKCYECGEPGHFARECRNRG 389
Score = 26.6 bits (57), Expect = 4.9
Identities = 8/21 (38%), Positives = 12/21 (57%)
Frame = +3
Query: 37 RCFKCNREGHLAVNCKTLQGG 57
+C++C GH A C+ GG
Sbjct: 333 KCYECGEPGHFARECRNRGGG 395
>AJ388976 similar to PIR|E84638|E84 probable RSZp22 splicing factor
[imported] - Arabidopsis thaliana, partial (62%)
Length = 508
Score = 31.6 bits (70), Expect = 0.15
Identities = 10/21 (47%), Positives = 14/21 (66%)
Frame = +2
Query: 123 CFNCNQPGHFSRDCKAPKGES 143
C+ C +PGHF+R C + G S
Sbjct: 344 CYXCGEPGHFARXCNSSPGGS 406
Score = 29.6 bits (65), Expect = 0.58
Identities = 17/51 (33%), Positives = 22/51 (42%), Gaps = 1/51 (1%)
Frame = +2
Query: 47 LAVNCKTLQGGPSDSKMKGNDATGGTGNKTSG-KIVTCYRCKKVGHYTNKC 96
L+ N KT GG + G GG G G + CY C + GH+ C
Sbjct: 239 LSHNSKT--GGGGGGRGGGGGGGGGRGRSGGGGSDLKCYXCGEPGHFARXC 385
Score = 27.3 bits (59), Expect = 2.9
Identities = 13/40 (32%), Positives = 17/40 (42%), Gaps = 6/40 (15%)
Frame = +2
Query: 37 RCFKCNREGHLAVNCKTLQGG------PSDSKMKGNDATG 70
+C+ C GH A C + GG PS + N TG
Sbjct: 341 KCYXCGEPGHFARXCNSSPGGSGXRRNPSPPQFXRNPITG 460
Score = 26.2 bits (56), Expect = 6.4
Identities = 8/14 (57%), Positives = 10/14 (71%)
Frame = +2
Query: 103 CFNCGEIGHYASKC 116
C+ CGE GH+A C
Sbjct: 344 CYXCGEPGHFARXC 385
>BG450974 similar to PIR|T05112|T05 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (54%)
Length = 364
Score = 31.2 bits (69), Expect = 0.20
Identities = 8/15 (53%), Positives = 13/15 (86%)
Frame = +1
Query: 123 CFNCNQPGHFSRDCK 137
C+ C +PGHF+R+C+
Sbjct: 319 CYECGEPGHFARECR 363
Score = 30.4 bits (67), Expect = 0.34
Identities = 12/37 (32%), Positives = 17/37 (45%)
Frame = +1
Query: 60 DSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKC 96
+SK G GG G + CY C + GH+ +C
Sbjct: 250 NSKSGGGGGRGGGRGGRGGDDLKCYECGEPGHFAREC 360
Score = 28.1 bits (61), Expect = 1.7
Identities = 8/15 (53%), Positives = 12/15 (79%)
Frame = +1
Query: 103 CFNCGEIGHYASKCK 117
C+ CGE GH+A +C+
Sbjct: 319 CYECGEPGHFARECR 363
>TC80683 homologue to GP|8777424|dbj|BAA97014.1
gb|AAF56406.1~gene_id:K9P8.7~strong similarity to unknown
protein {Arabidopsis thaliana}, partial (16%)
Length = 1360
Score = 30.4 bits (67), Expect = 0.34
Identities = 9/17 (52%), Positives = 13/17 (75%)
Frame = +1
Query: 121 RLCFNCNQPGHFSRDCK 137
++C+ C + GH SRDCK
Sbjct: 1072 KICYKCKKVGHLSRDCK 1122
Score = 29.3 bits (64), Expect = 0.75
Identities = 9/16 (56%), Positives = 12/16 (74%)
Frame = +1
Query: 83 CYRCKKVGHYTNKCPD 98
CY+CKKVGH + C +
Sbjct: 1078 CYKCKKVGHLSRDCKE 1125
Score = 26.6 bits (57), Expect = 4.9
Identities = 8/15 (53%), Positives = 12/15 (79%)
Frame = +1
Query: 38 CFKCNREGHLAVNCK 52
C+KC + GHL+ +CK
Sbjct: 1078 CYKCKKVGHLSRDCK 1122
Score = 26.2 bits (56), Expect = 6.4
Identities = 17/70 (24%), Positives = 27/70 (38%), Gaps = 5/70 (7%)
Frame = +1
Query: 95 KCPDGGPL-----CFNCGEIGHYASKCKIKGRLCFNCNQPGHFSRDCKAPKGESSGNTGK 149
K D GP+ C+ C ++GH + CK QP + E + N
Sbjct: 1039 KKSDSGPIDAPKICYKCKKVGHLSRDCK---------EQPNDLLHSHATSEAEENPNM-N 1188
Query: 150 GKQLAAEERI 159
L+ E+R+
Sbjct: 1189 ASNLSLEDRV 1218
>TC89153 similar to GP|18855061|gb|AAL79753.1 putative RNA helicase {Oryza
sativa}, partial (3%)
Length = 737
Score = 30.0 bits (66), Expect = 0.44
Identities = 10/17 (58%), Positives = 12/17 (69%)
Frame = +3
Query: 120 GRLCFNCNQPGHFSRDC 136
G CF+C QPGH + DC
Sbjct: 138 GGACFSCGQPGHRASDC 188
Score = 28.5 bits (62), Expect = 1.3
Identities = 15/44 (34%), Positives = 20/44 (45%)
Frame = +3
Query: 73 GNKTSGKIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKC 116
G ++SG Y N+ G CF+CG+ GH AS C
Sbjct: 72 GRRSSG-----YSSSNRSSSPNRRGSYGGACFSCGQPGHRASDC 188
>TC76340 homologue to SP|Q9SM09|VATA_CITUN Vacuolar ATP synthase catalytic
subunit A (EC 3.6.3.14) (V-ATPase A subunit), complete
Length = 2417
Score = 29.6 bits (65), Expect = 0.58
Identities = 15/33 (45%), Positives = 17/33 (51%)
Frame = +3
Query: 30 NCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSK 62
NC G CF R+ HL NCKT + G SK
Sbjct: 1587 NCPACGKGCFS*RRQDHLR-NCKTFERGLPCSK 1682
>TC85971 similar to GP|19424073|gb|AAL87350.1 unknown protein {Arabidopsis
thaliana}, partial (77%)
Length = 1540
Score = 28.9 bits (63), Expect = 0.98
Identities = 21/70 (30%), Positives = 27/70 (38%), Gaps = 7/70 (10%)
Frame = +3
Query: 82 TCYRCKKVGHYTNKCPDGGPLCFNCGEIGH------YASKCKIKGRL-CFNCNQPGHFSR 134
TC ++ GHY P C+ G I H KC KG+L C C G
Sbjct: 690 TCNADEEPGHYKENQVTQCPSCYGRGLIAHKDGSDTLCVKCSGKGKLPCATCGSRGLL-- 863
Query: 135 DCKAPKGESS 144
+C+ G S
Sbjct: 864 NCETCNGSGS 893
>TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pectin
methylesterase {Arabidopsis thaliana}, partial (33%)
Length = 1289
Score = 28.9 bits (63), Expect = 0.98
Identities = 25/78 (32%), Positives = 32/78 (40%), Gaps = 7/78 (8%)
Frame = +3
Query: 49 VNCKTLQGGPSDSKMKGNDATGGTGNKT-------SGKIVTCYRCKKVGHYTNKCPDGGP 101
+N + P SK++ A TGNKT SG T Y K + HY N C G
Sbjct: 531 INFENTASFPIGSKVEQAVAVRITGNKTAFYNCTFSGVQDTLYDHKGL-HYFNNCTIKGS 707
Query: 102 LCFNCGEIGHYASKCKIK 119
+ F CG C I+
Sbjct: 708 VDFICGHGKSLYEGCTIR 761
>TC81162 weakly similar to PIR|T47673|T47673 hypothetical protein T26I12.220
- Arabidopsis thaliana, partial (9%)
Length = 756
Score = 28.9 bits (63), Expect = 0.98
Identities = 10/24 (41%), Positives = 14/24 (57%)
Frame = +2
Query: 117 KIKGRLCFNCNQPGHFSRDCKAPK 140
K K R+C+ C Q GH +C P+
Sbjct: 404 KRKNRMCYGCRQKGHNLSECPNPQ 475
Score = 28.5 bits (62), Expect = 1.3
Identities = 16/55 (29%), Positives = 25/55 (45%)
Frame = +2
Query: 44 EGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCPD 98
E AV G PS + + A+G N+ CY C++ GH ++CP+
Sbjct: 323 EKSFAVEQPGAGGKPSSVEQPSSVASGKRKNRM------CYGCRQKGHNLSECPN 469
>TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.2 (TTS-2.2)
{Homo sapiens}, partial (2%)
Length = 1522
Score = 28.5 bits (62), Expect = 1.3
Identities = 13/39 (33%), Positives = 20/39 (50%), Gaps = 6/39 (15%)
Frame = -1
Query: 38 CFKCNREGHLAVNC------KTLQGGPSDSKMKGNDATG 70
C+ C+ GH+A NC K + GP +K++ A G
Sbjct: 316 CYSCHERGHIARNCTNTSAAKKKKKGPKVAKVEPQKAEG 200
Score = 25.8 bits (55), Expect = 8.3
Identities = 7/16 (43%), Positives = 13/16 (80%)
Frame = -1
Query: 121 RLCFNCNQPGHFSRDC 136
R C++C++ GH +R+C
Sbjct: 322 RECYSCHERGHIARNC 275
>TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA helicase
{Oryza sativa}, partial (7%)
Length = 624
Score = 28.5 bits (62), Expect = 1.3
Identities = 11/17 (64%), Positives = 11/17 (64%)
Frame = +1
Query: 103 CFNCGEIGHYASKCKIK 119
CF CGE GH AS C K
Sbjct: 166 CFTCGESGHRASDCPNK 216
Score = 26.6 bits (57), Expect = 4.9
Identities = 8/20 (40%), Positives = 12/20 (60%)
Frame = +1
Query: 123 CFNCNQPGHFSRDCKAPKGE 142
CF C + GH + DC +G+
Sbjct: 166 CFTCGESGHRASDCPNKRGD 225
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.140 0.472
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,289,318
Number of Sequences: 36976
Number of extensions: 130788
Number of successful extensions: 767
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 624
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 722
length of query: 165
length of database: 9,014,727
effective HSP length: 89
effective length of query: 76
effective length of database: 5,723,863
effective search space: 435013588
effective search space used: 435013588
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0306.5


