

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0306.4
(968 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
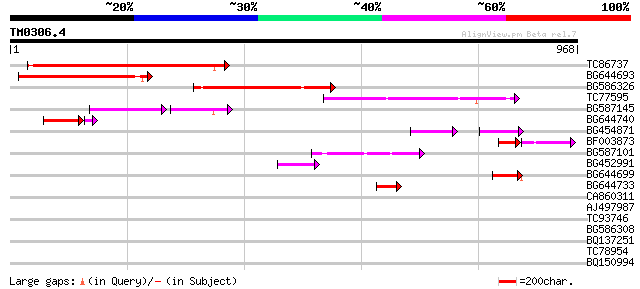
Score E
Sequences producing significant alignments: (bits) Value
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 282 4e-76
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 215 8e-56
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 211 8e-55
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 154 2e-37
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 95 3e-29
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 87 1e-18
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 65 2e-14
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 61 2e-13
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 50 3e-06
BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, pa... 46 8e-05
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 44 4e-04
BG644733 weakly similar to GP|15289942|db putative polyprotein {... 43 7e-04
CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product ... 41 0.002
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 38 0.016
TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical... 36 0.082
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 33 0.69
BQ137251 similar to GP|18026135|gb| cytochrome c oxidase subunit... 29 7.6
TC78954 weakly similar to PIR|D96810|D96810 hypothetical protein... 29 7.6
BQ150994 similar to GP|22122173|dbj preproneuropeptide B {Homo s... 29 10.0
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 282 bits (722), Expect = 4e-76
Identities = 156/352 (44%), Positives = 215/352 (60%), Gaps = 7/352 (1%)
Frame = +1
Query: 31 FGPISIAPYRMAPAELVELKSQLEDLSAKGFIRPSVSPWGAPVLLVKKKDGKSRLCVDYR 90
+GP+ Y M+ EL+ LK LEDL KGFI+ S S GAPVL V+K G R CVDYR
Sbjct: 469 WGPL----YGMSRQELLVLKKTLEDLLDKGFIKASGSAAGAPVLFVRKPGGGIRFCVDYR 636
Query: 91 QLNKATVKNRYPLPRIDDLMDQLRGDAVFSKIDLKSSYHQIRVKTEDIPKTAFRTRYGHY 150
LN T K+RYPLP I + + ++ G F+K+D+ +++H++R+K ED KTAFRTRYG +
Sbjct: 637 ALNAITKKDRYPLPLISETLRRVAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLF 816
Query: 151 EYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILIYS*DT-KEHEEHLCQVLHV 209
E++V PFG+T APA F Y+N+ HEFLD FV +IDD+LIY+ + K+HE + +VL
Sbjct: 817 EWIVCPFGLTGAPATFQRYINKTLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRR 996
Query: 210 LREKKLYANPSKCEFWLEEVNFLGHVISK-EGIVVDPAKVETVLAWEQPKTVTEIRSFVG 268
L + L +P KCEF + V ++G +++ +G+ DP K+ + W P +V RSF+G
Sbjct: 997 LADAGLSLDPKKCEFSVTTVKYVGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLG 1176
Query: 269 LAGYYRRFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQTLKERLTTSPVLILPQPEEP 328
YY+ FI G+++I PLT+LTRKD PF W + +F LK PVL + PE
Sbjct: 1177FCNYYKDFIPGYSEITEPLTRLTRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPEAV 1356
Query: 329 YEVYCDASHQGLGCVLMQH-----WKVVAYASRQLKTHEKNYPTHDLELTAI 375
V D S LG VL Q VA+ S++L E NYP HD EL A+
Sbjct: 1357TTVETDCSGFALGGVLTQEDGTGAAHPVAFHSQRLSPAEYNYPIHDKELLAV 1512
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 215 bits (547), Expect = 8e-56
Identities = 120/235 (51%), Positives = 150/235 (63%), Gaps = 5/235 (2%)
Frame = +2
Query: 15 LPPVRDVEFVIDIVRGFGPISIAPYRMAPAELVELKSQLEDLSAKGFIRPSVSPWGAPVL 74
+PP ++F ID++ PI I YR+ P +L LK QL+DL KGFI+PS+ P G VL
Sbjct: 17 VPPEWKIDFGIDLLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVVVL 196
Query: 75 LVKKKDGKSRLCVDYRQLNKATVKNRYPLPRIDDLMDQLRGDAVFSKIDLKSSYHQIRVK 134
+KKKDG R+ +DY QLN +K +YPLP ID+L D L+G F KIDL+ HQ RV
Sbjct: 197 FLKKKDGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVI 376
Query: 135 TEDIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILIYS* 194
ED+PKTAFR RYGHYE LVM FG TN P FM+ MNR+F ++LD V+VF +DILIYS
Sbjct: 377 GEDVPKTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSK 556
Query: 195 DTKEHEEHLCQVLHVLREKKLYANPSKCEF----WLEEVNFLG-HVISKEGIVVD 244
+ EHE HL L VL++ L C+ L EV F HVIS EG+ VD
Sbjct: 557 NENEHENHLRLALKVLKDIGL------CQISYV*ILVEVGFFSLHVISGEGLKVD 703
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 211 bits (538), Expect = 8e-55
Identities = 117/243 (48%), Positives = 154/243 (63%), Gaps = 2/243 (0%)
Frame = +2
Query: 315 TTSPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASRQLKTHEKNYPTHDLELTA 374
T++P+L+LP+ Y VY DAS GLGCVL QH KV+AYASRQL+ HE NYPTHDLE+ A
Sbjct: 8 TSAPILVLPELIT-YVVYTDASITGLGCVLTQHEKVIAYASRQLRKHEGNYPTHDLEMAA 184
Query: 375 IVFSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRWMEFIKDYEFTLQYHPGKA 434
+VF+LKIWR YLYG I +DHKSLKY+F Q ELN+RQRRWMEF+ DY+ + Y+PGKA
Sbjct: 185 VVFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDITYYPGKA 364
Query: 435 NVVADALSRKMHVSLKMVKELELLE-QFRDMSLGVTPYEGMLKFGMIRI-ASGLMEEIRE 492
N+VADALSR+ V + +E + L+ R + L V + G+ + + L IR
Sbjct: 365 NLVADALSRR-RVDVSAEREADDLDGMVRALRLNVLT-KATESLGLEAVNQADLFTRIRL 538
Query: 493 RQLQDVFLLEKRNLVIQGKDPEFKIGTDNILRCKDRVCVPTNPELRRLIMDEGHKSKWRI 552
Q QD E V Q E++ D + R+ VP + L+ IM E HKS++ +
Sbjct: 539 AQGQD----ENLQKVAQNDRTEYQTAKDGTILVNGRISVPNDRSLKEEIMSEAHKSRFSV 706
Query: 553 HPG 555
HPG
Sbjct: 707 HPG 715
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial
(14%)
Length = 1708
Score = 154 bits (389), Expect = 2e-37
Identities = 108/341 (31%), Positives = 168/341 (48%), Gaps = 7/341 (2%)
Frame = +2
Query: 536 ELRRLIMDEGHKSKWRIHPGMTKMYQDLKLNFWWPGMKKQVVEYVAACLTCQKAKVEHQR 595
ELR ++ E H S HPG + + F+WPG + V +V C C + Q
Sbjct: 164 ELRTKLVQESHDSTAAGHPGRNGTLEIVSRKFFWPGQSQTVRRFVRNCDVCGGIHIWRQA 343
Query: 596 PAGMLQSLDVPEWKWDSISMDFVVALPRTQKRHDS-IWVIVDRLTKSAHFLPGRTTYNVE 654
G L+ L VP +SMDF+ +LP T+ R +WVIVDRL+KS L T E
Sbjct: 344 KRGFLKPLPVPNRLHSDLSMDFITSLPPTRGRGSQYLWVIVDRLSKSVT-LEEMDTMEAE 520
Query: 655 KLAEIYVAEIVRLHGVSTSIVSDSDPKFTSHFWGALHDALGTKLRLSSAYHPQIDGQTER 714
A+ +++ R HG+ SIVSD + FW G LS++YHPQ DG TER
Sbjct: 521 ACAQRFLSCHYRFHGMPQSIVSDRGSNWVGRFWREFCRLTGVTQLLSTSYHPQTDGGTER 700
Query: 715 TIQSLEDLLRACVLDNRGSWDDLQPLIEFTYNNNFHASIGMAPYEALYGRKCRTPLCWYQ 774
Q ++ +LRA V ++ +W DL P ++ N ++SIG P+ +G P+ +
Sbjct: 701 WNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSIGATPFFVEHGYHV-DPIPTVE 877
Query: 775 D-GESLLIGPELLQQTTEKVKQ----IREKMRASQSRQKSYADQRRRTLE-FEEGDHVFL 828
D G + G Q +++K I+ ++ A+Q R ++ A++RR + ++ GD V+L
Sbjct: 878 DTGGVVSEGEAAAQLLVKRMKDVTGFIQAEIVAAQQRSEASANKRRCPADRYQVGDKVWL 1057
Query: 829 RVTQTTGVGRAIKSRKLTPKFIGPYQITRRVGPVAYQIALP 869
V+ + K L K Y++TR V P ++ +P
Sbjct: 1058NVSNYKSPRPSKKLDWLHHK----YEVTRFVTPHVVELNVP 1168
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 95.1 bits (235), Expect(2) = 3e-29
Identities = 48/133 (36%), Positives = 77/133 (57%)
Frame = +2
Query: 136 EDIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILIYS*D 195
+D+ KTAF T G Y Y VMPFG+ NA + + +NR+F + L + V+IDD+L+ S
Sbjct: 11 DDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVKSLR 190
Query: 196 TKEHEEHLCQVLHVLREKKLYANPSKCEFWLEEVNFLGHVISKEGIVVDPAKVETVLAWE 255
+H HL + L E + NP+KC F + FLG++++++GI V+P ++ +L
Sbjct: 191 ATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAILDLP 370
Query: 256 QPKTVTEIRSFVG 268
PK E++ G
Sbjct: 371 SPKNSREVQRLTG 409
Score = 52.8 bits (125), Expect(2) = 3e-29
Identities = 32/110 (29%), Positives = 53/110 (48%), Gaps = 4/110 (3%)
Frame = +3
Query: 275 RFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQTLKERLTTSPVLILPQPEEPYEVYCD 334
RFI P +L ++ F W E C+ +F+ LK+ LTT PVL P+ + +Y
Sbjct: 429 RFISRSTDKCLPFYKLLCGNKRFVWDEKCEEAFEQLKQYLTTPPVLSKPEAGDTLSLYIA 608
Query: 335 ASHQGLGCVLMQ----HWKVVAYASRQLKTHEKNYPTHDLELTAIVFSLK 380
S + VL++ K + Y S+++ E YPT + A++ S +
Sbjct: 609 ISSTAVSSVLIREDRGEQKPIFYTSKRMTDPETRYPTLEKMAFAVITSAR 758
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 86.7 bits (213), Expect(2) = 1e-18
Identities = 39/67 (58%), Positives = 51/67 (75%)
Frame = -1
Query: 59 KGFIRPSVSPWGAPVLLVKKKDGKSRLCVDYRQLNKATVKNRYPLPRIDDLMDQLRGDAV 118
K F +PS+SP GA +L V+KKDG R+C+DYRQ NK T KN+YPLPRID+L D+++ D
Sbjct: 274 KRFQQPSISP*GAALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCY 95
Query: 119 FSKIDLK 125
F IDL+
Sbjct: 94 F*NIDLR 74
Score = 25.4 bits (54), Expect(2) = 1e-18
Identities = 11/22 (50%), Positives = 13/22 (59%)
Frame = -3
Query: 128 YHQIRVKTEDIPKTAFRTRYGH 149
YHQ RV +IPKT T G+
Sbjct: 68 YHQFRVNEVNIPKTTLLTWRGY 3
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 65.5 bits (158), Expect(2) = 2e-14
Identities = 33/81 (40%), Positives = 45/81 (54%)
Frame = +2
Query: 684 SHFWGALHDALGTKLRLSSAYHPQIDGQTERTIQSLEDLLRACVLDNRGSWDDLQPLIEF 743
S+FW L GT L +SSAYHP DGQ+E + E LR + + W P E+
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 744 TYNNNFHASIGMAPYEALYGR 764
YN +++ S M P++ALYGR
Sbjct: 212 WYNTSYNISAAMTPFKALYGR 274
Score = 32.7 bits (73), Expect(2) = 2e-14
Identities = 21/77 (27%), Positives = 36/77 (46%), Gaps = 2/77 (2%)
Frame = +1
Query: 803 SQSRQKSYADQRRRTLEFEEGDHVFLRVT--QTTGVGRAIKSRKLTPKFIGPYQITRRVG 860
+Q K AD++RR EF+ G+HV +++ Q + V + +P F +
Sbjct: 388 AQQTMKHQADKKRRHFEFQLGEHVLVKLQPYQQSSVALRKYQKFGSPNFGSLLTVCSL*V 567
Query: 861 PVAYQIALPPFLPIFTM 877
A+ PP+L FT+
Sbjct: 568 ESAFHCKSPPYLSCFTI 618
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 60.8 bits (146), Expect(2) = 2e-13
Identities = 27/38 (71%), Positives = 33/38 (86%)
Frame = +2
Query: 835 GVGRAIKSRKLTPKFIGPYQITRRVGPVAYQIALPPFL 872
GVGRA+KS+KLT +FIGPYQI+ RVG VAY++ LPP L
Sbjct: 2 GVGRALKSKKLTVRFIGPYQISERVGTVAYRVGLPPHL 115
Score = 33.9 bits (76), Expect(2) = 2e-13
Identities = 34/96 (35%), Positives = 48/96 (49%), Gaps = 3/96 (3%)
Frame = +3
Query: 874 IFTMYFMCRS*GSTFQTRHM*SSQTTLNCRMISLLRHR--RSALGTEE*NS*EGSRLH** 931
I M FMCR+ GS F+ M*S + C+ + LR R R L E+* *E R *
Sbjct: 120 ICMMSFMCRNFGSMFRIHLM*S--RVMMCKSETTLR*RLYR*GLMIEK*RH*EARRYLL* 293
Query: 932 KCYGT-TIRETLHGNWKTRSRNCIPDFLRKPEFRGR 966
+ +GT + + G+ + R + I L + FRGR
Sbjct: 294 ESFGTERMVKA*RGSLRVRWWSLIQSCLHEVNFRGR 401
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 50.4 bits (119), Expect = 3e-06
Identities = 46/198 (23%), Positives = 87/198 (43%), Gaps = 5/198 (2%)
Frame = +2
Query: 515 FKIGTDNI-LRCKDRVCVPTNPELRRLIMDEGHKSKWRIHPGMTKMYQDLK-LNFWWPGM 572
+K DNI +RC +P I+ H S + H ++K ++ FWWP M
Sbjct: 44 YKQCADNIYIRCVAEEEIPG-------ILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTM 202
Query: 573 KKQVVEYVAACLTCQK---AKVEHQRPAGMLQSLDVPEWKWDSISMDFVVALPRTQKRHD 629
K +++ C CQ+ ++ P + ++V +D +DF+ P + +
Sbjct: 203 FKDAHSFISKCDPCQRQGNIS*RNEMPQNFILEVEV----FDVWGIDFMGPFP-SSYNNK 367
Query: 630 SIWVIVDRLTKSAHFLPGRTTYNVEKLAEIYVAEIVRLHGVSTSIVSDSDPKFTSHFWGA 689
I V VD ++K + T + + +++ + I GV ++SD F + +
Sbjct: 368 YILVAVDYVSKWVEAIASPTN-DATVVVKMFKSVIFPRFGVPRVVISDGGSHFINKVFEK 544
Query: 690 LHDALGTKLRLSSAYHPQ 707
L G + ++++AYHPQ
Sbjct: 545 LLKKNGVRHKVATAYHPQ 598
>BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, partial (33%)
Length = 560
Score = 45.8 bits (107), Expect = 8e-05
Identities = 25/72 (34%), Positives = 42/72 (57%)
Frame = +3
Query: 458 LEQFRDMSLGVTPYEGMLKFGMIRIASGLMEEIRERQLQDVFLLEKRNLVIQGKDPEFKI 517
LEQFRD+SL +K GM++I + ++ I+E Q DV L++ Q +D +FK+
Sbjct: 69 LEQFRDLSLVCEVSPQSVKLGMLKINNEFLDSIKEAQKVDVKLVDLMFGNNQTEDGDFKV 248
Query: 518 GTDNILRCKDRV 529
+L+ +DR+
Sbjct: 249 DDQGVLQFRDRI 284
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 43.5 bits (101), Expect = 4e-04
Identities = 24/57 (42%), Positives = 37/57 (64%), Gaps = 5/57 (8%)
Frame = +2
Query: 824 DHVFLRVTQTT-GVGRAIKSRKLTPKFIGPYQITRRVGPVAYQIALPPFL----PIF 875
+ V L+V T G R K KL+ ++IGP+++ +R+G VAY++ALPP L P+F
Sbjct: 2 EQVLLKVLPTERGDCRFGKRGKLSLRYIGPFEVIKRIGEVAYELALPPGLSGVHPVF 172
>BG644733 weakly similar to GP|15289942|db putative polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (1%)
Length = 174
Score = 42.7 bits (99), Expect = 7e-04
Identities = 21/43 (48%), Positives = 31/43 (71%)
Frame = -3
Query: 626 KRHDSIWVIVDRLTKSAHFLPGRTTYNVEKLAEIYVAEIVRLH 668
K ++SI V+VDRLTKS F+P +T+Y+ + A I + EIV +H
Sbjct: 130 KSYESI*VVVDRLTKSTLFIPFKTSYSAK*YARILLDEIVCIH 2
>CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product {Drosophila
melanogaster}, partial (20%)
Length = 192
Score = 41.2 bits (95), Expect = 0.002
Identities = 22/60 (36%), Positives = 34/60 (56%), Gaps = 5/60 (8%)
Frame = +1
Query: 339 GLGCVLMQ-----HWKVVAYASRQLKTHEKNYPTHDLELTAIVFSLKIWRHYLYGCTFTI 393
G+G L Q H +AYASR L E+NY + E A +++++ +RHYL+G F +
Sbjct: 13 GIGAGLSQKDEENHEHPIAYASRLLTAAERNYTVVERECLAAIWAIRNFRHYLHGPKFEL 192
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 38.1 bits (87), Expect = 0.016
Identities = 21/74 (28%), Positives = 40/74 (53%), Gaps = 3/74 (4%)
Frame = -2
Query: 374 AIVFSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRWMEFIKDYEFTLQYHPGK 433
A+ ++ K RHY+ T + S +KY+F++ L R RW + +Y+ ++Y K
Sbjct: 623 ALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYD--IEYRSQK 450
Query: 434 A---NVVADALSRK 444
A +++AD L+ +
Sbjct: 449 AIKGSILADHLAHQ 408
>TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical
protein~similar to gag-pol polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (4%)
Length = 1019
Score = 35.8 bits (81), Expect = 0.082
Identities = 15/41 (36%), Positives = 27/41 (65%)
Frame = +2
Query: 605 VPEWKWDSISMDFVVALPRTQKRHDSIWVIVDRLTKSAHFL 645
VP+ W+ +++DF + L TQ+ DS V+ D+ ++ AHF+
Sbjct: 476 VPKPPWEDVTIDFSLGLL*TQQLKDSKMVVGDKFSRMAHFI 598
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 686
Score = 32.7 bits (73), Expect = 0.69
Identities = 19/69 (27%), Positives = 35/69 (50%)
Frame = -2
Query: 668 HGVSTSIVSDSDPKFTSHFWGALHDALGTKLRLSSAYHPQIDGQTERTIQSLEDLLRACV 727
HG+ IV+D+ F S+ + + +L +S +PQ +GQ E + + + D L+ +
Sbjct: 685 HGLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKIIIDGLKKRL 506
Query: 728 LDNRGSWDD 736
+G W D
Sbjct: 505 DLKKGCWAD 479
>BQ137251 similar to GP|18026135|gb| cytochrome c oxidase subunit II
{Sphaerospira fraseri}, partial (12%)
Length = 1073
Score = 29.3 bits (64), Expect = 7.6
Identities = 21/63 (33%), Positives = 27/63 (42%)
Frame = -3
Query: 699 RLSSAYHPQIDGQTERTIQSLEDLLRACVLDNRGSWDDLQPLIEFTYNNNFHASIGMAPY 758
RLS + P D R + L LLR SW PL+EF ++ A G AP
Sbjct: 300 RLSCPFRPPPDTGL-RNLSFLPPLLRNFCFSVSLSWRQPSPLVEFRWSPRCAARAGRAPV 124
Query: 759 EAL 761
+L
Sbjct: 123 VSL 115
>TC78954 weakly similar to PIR|D96810|D96810 hypothetical protein T11I11.6
[imported] - Arabidopsis thaliana, partial (45%)
Length = 2329
Score = 29.3 bits (64), Expect = 7.6
Identities = 20/67 (29%), Positives = 34/67 (49%)
Frame = +1
Query: 401 KYLFDQKELNMRQRRWMEFIKDYEFTLQYHPGKANVVADALSRKMHVSLKMVKELELLEQ 460
K + + + N + RW I+DYE ++ PG VA AL ++ + LKM++ E
Sbjct: 1711 KAMLRRADCNAKLERWEAAIQDYEMLIREKPGDEE-VARALF-EIQLKLKMLRG----ED 1872
Query: 461 FRDMSLG 467
+D+ G
Sbjct: 1873 VKDLKFG 1893
>BQ150994 similar to GP|22122173|dbj preproneuropeptide B {Homo sapiens},
partial (15%)
Length = 1036
Score = 28.9 bits (63), Expect = 10.0
Identities = 19/63 (30%), Positives = 31/63 (49%)
Frame = +1
Query: 788 QTTEKVKQIREKMRASQSRQKSYADQRRRTLEFEEGDHVFLRVTQTTGVGRAIKSRKLTP 847
QTTEK Q REK RA + R+ ++ ++ T ++ H R T++ R ++ P
Sbjct: 526 QTTEKKDQTREKQRA-EKRRDGRSETQQSTQHAQDPRHPRTRATRSEEGPRGTRTEDNQP 702
Query: 848 KFI 850
I
Sbjct: 703 PTI 711
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.328 0.141 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,497,749
Number of Sequences: 36976
Number of extensions: 432911
Number of successful extensions: 2416
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 1787
Number of HSP's successfully gapped in prelim test: 85
Number of HSP's that attempted gapping in prelim test: 594
Number of HSP's gapped (non-prelim): 1918
length of query: 968
length of database: 9,014,727
effective HSP length: 105
effective length of query: 863
effective length of database: 5,132,247
effective search space: 4429129161
effective search space used: 4429129161
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0306.4


