

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0306.1
(1224 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
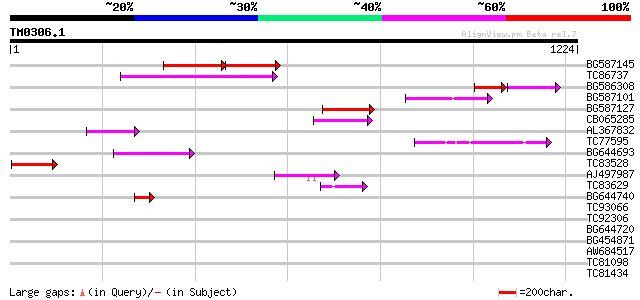
Score E
Sequences producing significant alignments: (bits) Value
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 155 7e-65
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 151 2e-36
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 81 1e-26
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 118 2e-26
BG587127 weakly similar to PIR|H84506|H84 probable retroelement ... 95 2e-19
CB065285 weakly similar to PIR|A84500|A84 probable retroelement ... 92 2e-18
AL367832 weakly similar to GP|14091845|gb Putative retroelement ... 89 8e-18
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 85 1e-16
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 85 1e-16
TC83528 80 6e-15
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 66 7e-11
TC83629 weakly similar to GP|6692261|gb|AAF24611.1| putative RNa... 50 4e-06
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 47 6e-05
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 39 0.009
TC92306 39 0.016
BG644720 34 0.30
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 33 0.52
AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thalia... 32 1.2
TC81098 homologue to GP|20373023|dbj|BAB91180. lipoic acid synth... 30 5.7
TC81434 24 8.8
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 155 bits (392), Expect(2) = 7e-65
Identities = 71/136 (52%), Positives = 103/136 (75%)
Frame = +2
Query: 332 MYAPDEEKTAFMTNQANYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEIYVDDMVVK 391
M+ D EKTAF+T++ YCY+ MPFGLKNAG+TYQRL++R+F ++G ME+Y+DDM+VK
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 392 SEEMGGHCLDLAEAFGEIRKHNMRLNPEKCSFGIQSGKFLGFMITRRGIEVNPDKCKAIL 451
S H L E F + ++ M+LNP KC+FG+ SG+FLG+++T++GIEVNP + AIL
Sbjct: 182 SLRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAIL 361
Query: 452 EMQSPTSVKEVQKLIG 467
++ SP + +EVQ+L G
Sbjct: 362 DLPSPKNSREVQRLTG 409
Score = 112 bits (279), Expect(2) = 7e-65
Identities = 56/118 (47%), Positives = 81/118 (68%)
Frame = +3
Query: 467 GRIAALSRFLPCSGSKATPFFQCLRKNRVFQWTDECEQAFQSLKELLSKPPILSRPIPGT 526
GRIAAL+RF+ S K PF++ L N+ F W ++CE+AF+ LK+ L+ PP+LS+P G
Sbjct: 408 GRIAALNRFISRSTDKCLPFYKLLCGNKRFVWDEKCEEAFEQLKQYLTTPPVLSKPEAGD 587
Query: 527 PLSVFISISDNAVSSVLLQECKDELRIIYFVSHALQGAELRYQKIEKAALALIISARK 584
LS++I+IS AVSSVL++E + E + I++ S + E RY +EK A A+I SARK
Sbjct: 588 TLSLYIAISSTAVSSVLIREDRGEQKPIFYTSKRMTDPETRYPTLEKMAFAVITSARK 761
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 151 bits (381), Expect = 2e-36
Identities = 102/343 (29%), Positives = 162/343 (46%), Gaps = 3/343 (0%)
Frame = +1
Query: 239 MGEEKAQAVKAETNKLIDAGFIREVKYPTWLANVVMVKKSNGKWRMCTDYTDLNKHCPKD 298
M ++ +K L+D GFI+ A V+ V+K G R C DY LN KD
Sbjct: 487 MSRQELLVLKKTLEDLLDKGFIKASGSAAG-APVLFVRKPGGGIRFCVDYRALNAITKKD 663
Query: 299 SYPLPNIDKLVDRASGFGMLSLMDAYSGYHQIRMYAPDEEKTAFMTNQANYCYQTMPFGL 358
YPLP I + + R +G + +D + +H++R+ D+EKTAF T + + PFGL
Sbjct: 664 RYPLPLISETLRRVAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVCPFGL 843
Query: 359 KNAGATYQRLMDRVFEGQVGRNMEIYVDDMVV-KSEEMGGHCLDLAEAFGEIRKHNMRLN 417
A AT+QR +++ + + Y+DD+++ + H + + + L+
Sbjct: 844 TGAPATFQRYINKTLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRRLADAGLSLD 1023
Query: 418 PEKCSFGIQSGKFLGFMITR-RGIEVNPDKCKAILEMQSPTSVKEVQKLIGRIAALSRFL 476
P+KC F + + K++GF++T +G+ +P K AI + P SVK + +G F+
Sbjct: 1024PKKCEFSVTTVKYVGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLGFCNYYKDFI 1203
Query: 477 PCSGSKATPFFQCLRKNRVFQWTDECEQAFQSLKELLSKPPILSRPIPGTPLSVFISISD 536
P P + RK+ F+W E E AF LK L ++ P+L P +V S
Sbjct: 1204PGYSEITEPLTRLTRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPEAVTTVETDCSG 1383
Query: 537 NAVSSVLLQE-CKDELRIIYFVSHALQGAELRYQKIEKAALAL 578
A+ VL QE + F S L AE Y +K LA+
Sbjct: 1384FALGGVLTQEDGTGAAHPVAFHSQRLSPAEYNYPIHDKELLAV 1512
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 686
Score = 80.9 bits (198), Expect(2) = 1e-26
Identities = 37/70 (52%), Positives = 49/70 (69%)
Frame = -2
Query: 1003 GVPATLVSDNGTQFTSRIVRDFCNEMGIEMRFASVEHPQSNGQVEAANKVILNGIKKRLG 1062
G+P +V+DNG+ F S R+FC I + AS +PQSNGQ EA+NK+I++G+KKRL
Sbjct: 682 GLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKIIIDGLKKRLD 503
Query: 1063 DAKGLWADEL 1072
KG WADEL
Sbjct: 502 LKKGCWADEL 473
Score = 58.9 bits (141), Expect(2) = 1e-26
Identities = 37/119 (31%), Positives = 62/119 (52%), Gaps = 5/119 (4%)
Frame = -3
Query: 1075 VVWAYNTTPQSTTGETPFRLTYGVDAMVPVEIQDMTFRVAAYDEN-ENHENRLID-LNLA 1132
V+W++ T P+ T TPF + + V+AM P E+ + + +N E + +RL + L
Sbjct: 465 VLWSHRTNPRGATKSTPFSMAHRVEAMAPAEVNVTSL*RSRMPQNIELNNDRLFNALETI 286
Query: 1133 EEVKTEVRLRQAAVKQRSERRYNTRVVPRHMQVGDLVLRR---KAKGPDDSKLSPNWEG 1188
EE + + LR + + E YN V + +++GD+VL + K + KL NWEG
Sbjct: 285 EERRDQALLRIQNYQHQIESYYNKTVKSQPLKLGDIVLCKVFENTKELNAGKLGTNWEG 109
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 118 bits (295), Expect = 2e-26
Identities = 59/187 (31%), Positives = 96/187 (50%)
Frame = +2
Query: 855 LYRRGVGVPLLRCVTRDEADRIMFEVHEGVCASHVGGRSLAAKVLRAGFYWPTLKNDCMG 914
LY++ +RCV +E I+F H A H +K+ +AGF+WPT+ D
Sbjct: 41 LYKQCADNIYIRCVAEEEIPGILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHS 220
Query: 915 YAKKCEKCQIYADLHRAPPEVLSSMSSAWPFAMWGVDILGPFTPAGAQIRFVLVAVDYFT 974
+ KC+ CQ ++ + + F +WG+D +GPF P+ +++LVAVDY +
Sbjct: 221 FISKCDPCQRQGNIS*RNEMPQNFILEVEVFDVWGIDFMGPF-PSSYNNKYILVAVDYVS 397
Query: 975 KWIEAESMAKITAEKVKKFYWRKIICRFGVPATLVSDNGTQFTSRIVRDFCNEMGIEMRF 1034
KW+EA + A V K + I RFGVP ++SD G+ F +++ + G+ +
Sbjct: 398 KWVEAIASPTNDATVVVKMFKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHKV 577
Query: 1035 ASVEHPQ 1041
A+ HPQ
Sbjct: 578 ATAYHPQ 598
>BG587127 weakly similar to PIR|H84506|H84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(13%)
Length = 415
Score = 94.7 bits (234), Expect = 2e-19
Identities = 46/113 (40%), Positives = 70/113 (61%)
Frame = +3
Query: 675 GVVLEGPGGVIIEQSLKFDFKASNNQAEYEAIIAGINLAIEMNVHCLVIKTDSQLVANQI 734
G+ L P I++QS + +F ASNN+ YEA+IAG+ LA + + + DSQLVA+Q
Sbjct: 3 GIRLTSPTNEILKQSFRLEFHASNNETNYEALIAGVRLAHGLKIRNIHAYCDSQLVASQF 182
Query: 735 KGDYQAKDIQLAKYLTKTQELMKRMDSVQVNHVPREENTRADVLCKLASTKKP 787
G+Y+A+D + YL Q+L +++D + +PR EN +AD L LAS+ P
Sbjct: 183 SGEYEARDELMDTYLKLVQKLAQKLDYFALTRIPRSENVQADALAALASSSDP 341
>CB065285 weakly similar to PIR|A84500|A84 probable retroelement gag/pol
polyprotein [imported] - Arabidopsis thaliana, partial
(2%)
Length = 592
Score = 91.7 bits (226), Expect = 2e-18
Identities = 50/127 (39%), Positives = 71/127 (55%)
Frame = -2
Query: 657 AEWILSVDGSSYLKGSGAGVVLEGPGGVIIEQSLKFDFKASNNQAEYEAIIAGINLAIEM 716
++W L DG+ G G G V+ P G I + + F+ +NN AEYEA I GI AI+M
Sbjct: 540 SKWGLVFDGAVNAYGKGIGAVIVSPQGHYIPFTARILFECTNNMAEYEACIFGIEEAIDM 361
Query: 717 NVHCLVIKTDSQLVANQIKGDYQAKDIQLAKYLTKTQELMKRMDSVQVNHVPREENTRAD 776
+ L I DS LV NQIKG+++ L Y + L+ V+++H+PR+EN AD
Sbjct: 360 RIKHLDIYGDSALVINQIKGEWETHHANLIPYRDYARRLLTYFTKVELHHIPRDENQMAD 181
Query: 777 VLCKLAS 783
L L+S
Sbjct: 180 ALATLSS 160
>AL367832 weakly similar to GP|14091845|gb Putative retroelement {Oryza
sativa}, partial (2%)
Length = 384
Score = 89.4 bits (220), Expect = 8e-18
Identities = 44/119 (36%), Positives = 71/119 (58%), Gaps = 4/119 (3%)
Frame = -1
Query: 166 EEETKTVKVG----ERNLKVGVNLTAIQEARLTQLLAENMDLFAWSAQDLPGIDPNFICH 221
+EE + + +G +R +KVG L + ++ QLL E +D+FA S +D+PG+DP + H
Sbjct: 357 QEEIELINLGTEENKREIKVGAALEEGVKRKIFQLLREYLDIFACSYEDMPGLDPKIVEH 178
Query: 222 KLALNPGVKPIAQMKRKMGEEKAQAVKAETNKLIDAGFIREVKYPTWLANVVMVKKSNG 280
++ P P+ R+ + A +K+E K IDAGF+ V+YP W+AN+V V K +G
Sbjct: 177 RIPTKPECPPVR*KLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDG 1
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 85.1 bits (209), Expect = 1e-16
Identities = 82/307 (26%), Positives = 135/307 (43%), Gaps = 12/307 (3%)
Frame = +2
Query: 875 RIMFEVHEGVCASHVGGRSLAAKVLRAGFYWPTLKNDCMGYAKKCEKC---QIYADLHRA 931
+++ E H+ A H G R+ +++ F+WP + + C+ C I+ R
Sbjct: 176 KLVQESHDSTAAGHPG-RNGTLEIVSRKFFWPGQSQTVRRFVRNCDVCGGIHIWRQAKRG 352
Query: 932 ---PPEVLSSMSSAWPFAMWGVDILGPFTPAGAQIRFVLVAVDYFTKWIEAESMAKITAE 988
P V + + S +M + L P G+Q ++ V VD +K + E M + AE
Sbjct: 353 FLKPLPVPNRLHS--DLSMDFITSLPPTRGRGSQ--YLWVIVDRLSKSVTLEEMDTMEAE 520
Query: 989 KVKKFYWRKIICRF---GVPATLVSDNGTQFTSRIVRDFCNEMGIEMRFASVEHPQSNGQ 1045
+ R + C + G+P ++VSD G+ + R R+FC G+ ++ HPQ++G
Sbjct: 521 ACAQ---RFLSCHYRFHGMPQSIVSDRGSNWVGRFWREFCRLTGVTQLLSTSYHPQTDGG 691
Query: 1046 VEAANKVILNGIKKRLGDAKGLWADELLTVVWAYNTTPQSTTGETPFRLTYG--VDAMVP 1103
E N+ I ++ + ++ W D L TV A S+ G TPF + +G VD +
Sbjct: 692 TERWNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSIGATPFFVEHGYHVDPIPT 871
Query: 1104 VEIQDMTFRVAAYDENENHENRLIDLNLAEEVKTEVRLRQAAVKQRSERRYNTRVVPR-H 1162
VE E E L+ ++V ++ A +QRSE N R P
Sbjct: 872 VEDTG-----GVVSEGEAAAQLLV--KRMKDVTGFIQAEIVAAQQRSEASANKRRCPADR 1030
Query: 1163 MQVGDLV 1169
QVGD V
Sbjct: 1031 YQVGDKV 1051
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 85.1 bits (209), Expect = 1e-16
Identities = 55/174 (31%), Positives = 84/174 (47%)
Frame = +2
Query: 225 LNPGVKPIAQMKRKMGEEKAQAVKAETNKLIDAGFIREVKYPTWLANVVMVKKSNGKWRM 284
L P + PI ++ K + +K + L++ GFI+ YP + V+ +KK +G RM
Sbjct: 53 LLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVV-VLFLKKKDGFLRM 229
Query: 285 CTDYTDLNKHCPKDSYPLPNIDKLVDRASGFGMLSLMDAYSGYHQIRMYAPDEEKTAFMT 344
DY LN K YPLP ID+L D G +D G HQ R+ D KTAF
Sbjct: 230 SIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVIGEDVPKTAFRI 409
Query: 345 NQANYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEIYVDDMVVKSEEMGGH 398
+Y M FG N + LM+RVF+ + + ++ +D+++ S+ H
Sbjct: 410 RYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSKNENEH 571
>TC83528
Length = 555
Score = 79.7 bits (195), Expect = 6e-15
Identities = 40/103 (38%), Positives = 64/103 (61%), Gaps = 3/103 (2%)
Frame = +3
Query: 4 SSADLIYGDAYEKLGLTEADLLPYDGALVGFSGERVFVRGYVELNTVFGEGINAESFAIK 63
S D++Y A+ K+ L E+ L P G L G G+ + V+GY++L+T FG+G N ++ ++
Sbjct: 246 SKVDILYWSAFLKMKLQESMLKPCQGFLKGTFGKGLPVKGYIDLDTTFGKGENTKTIKVR 425
Query: 64 FLVVK---CTSPYNVLIGRPSLNKLGAIISTRHLTVKYPLSKG 103
+ VV+ S YNV++G PSL L A++S T+KYP+ G
Sbjct: 426 YFVVESPPSVSIYNVVLGWPSLKDLKAVLSVAEFTIKYPVGDG 554
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 66.2 bits (160), Expect = 7e-11
Identities = 48/168 (28%), Positives = 77/168 (45%), Gaps = 29/168 (17%)
Frame = -2
Query: 573 KAALALIISARKLRPYFQGFQIKVKTDF-PLRQVLQKPDLAGRMVSWAVELSEFGIVFEK 631
K AL +A++LR Y + + P++ + +KP L GR+ W + LSE+ I +
Sbjct: 635 KTCCALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYDIEYRS 456
Query: 632 KGQVKAQVLAD----------------FVNEMSPEVKVSE------------EAEWILSV 663
+ +K +LAD F +E +K+ + ++ W L
Sbjct: 455 QKAIKGSILADHLAHQPLEDYRPIKFDFPDEEIMYLKMKDCDEPLFGEGPDPDSVWGLIF 276
Query: 664 DGSSYLKGSGAGVVLEGPGGVIIEQSLKFDFKASNNQAEYEAIIAGIN 711
DG+ + G+G G VL P G I + + F +NN AEYEA I GI+
Sbjct: 275 DGAVNVYGNGIGAVLLTPKGAHIPFTARLRFDCTNNIAEYEACIMGIH 132
>TC83629 weakly similar to GP|6692261|gb|AAF24611.1| putative RNase H
{Arabidopsis thaliana}, partial (21%)
Length = 1071
Score = 50.4 bits (119), Expect = 4e-06
Identities = 34/104 (32%), Positives = 49/104 (46%), Gaps = 3/104 (2%)
Frame = +2
Query: 672 SGAGVVLEGPGGVII---EQSLKFDFKASNNQAEYEAIIAGINLAIEMNVHCLVIKTDSQ 728
+GAG +L G ++ Q L K S AEY A++ G+ A + K DS+
Sbjct: 599 AGAGALLLAEDGSLLYGFRQGLGHQTKES---AEYRALLLGLKHASMKGFKYVTAKGDSE 769
Query: 729 LVANQIKGDYQAKDIQLAKYLTKTQELMKRMDSVQVNHVPREEN 772
LV NQI ++ KD L K + EL S ++ H+ RE N
Sbjct: 770 LVINQILDPWKIKDEHLKKLCAEALELSDNFHSFRIQHISRERN 901
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 46.6 bits (109), Expect = 6e-05
Identities = 20/42 (47%), Positives = 27/42 (63%)
Frame = -1
Query: 270 ANVVMVKKSNGKWRMCTDYTDLNKHCPKDSYPLPNIDKLVDR 311
A ++ V+K +G +RMC DY NK K+ YPLP ID L D+
Sbjct: 238 AALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDK 113
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 39.3 bits (90), Expect = 0.009
Identities = 38/146 (26%), Positives = 61/146 (41%), Gaps = 11/146 (7%)
Frame = +1
Query: 951 DILGP--FTPAGAQIRFVLVAVDYFTK--WIEAESMAKITAEKVKKFYWRKII-CRFGVP 1005
D+ GP T G + R+++ +D F + W+ T KK WR ++ + G
Sbjct: 115 DLWGPSKVTSYGGR-RYMMTIIDDFPRKVWVYFLRYKNETFPTFKK--WRILVETQTGKN 285
Query: 1006 AT-LVSDNGTQFTSRIVRDFCNEMGIEMRFASVEHPQSNGQVEAANKVILNGIKKRLGDA 1064
L++DN +F S +FC GI +PQ NG E + +L + L +A
Sbjct: 286 VKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCMLSNA 465
Query: 1065 -----KGLWADELLTVVWAYNTTPQS 1085
+ LW + T N +P S
Sbjct: 466 GL*N*RDLWVEAASTACHLVNRSPHS 543
>TC92306
Length = 521
Score = 38.5 bits (88), Expect = 0.016
Identities = 22/70 (31%), Positives = 29/70 (41%)
Frame = +2
Query: 707 IAGINLAIEMNVHCLVIKTDSQLVANQIKGDYQAKDIQLAKYLTKTQELMKRMDSVQVNH 766
I G+N A + ++ D Q V Q G ++ + L EL SV V H
Sbjct: 2 ILGLNEARNQGYEHVHVRGDFQXVCKQFXGSWKVNNPNLRNLCNXAVELKSNFKSVSVEH 181
Query: 767 VPREENTRAD 776
VPR N AD
Sbjct: 182 VPRGXNXAAD 211
>BG644720
Length = 678
Score = 34.3 bits (77), Expect = 0.30
Identities = 27/96 (28%), Positives = 40/96 (41%), Gaps = 3/96 (3%)
Frame = -3
Query: 864 LLRCVTRDEADRIMFEVHEGVCASHVGGRSLAAKVLRAGFYWPT---LKNDCMGYAKKCE 920
LL C+ +E + H VC SH L + R G+YW T LK
Sbjct: 325 LL*CLGEEETIQAFQ*AHSRVCGSHKS--KLYFHIKRMGYYWNT*IMLKG---------- 182
Query: 921 KCQIYADLHRAPPEVLSSMSSAWPFAMWGVDILGPF 956
+ A+ + PPEVL ++ F W +++ G F
Sbjct: 181 --ALLANFIQQPPEVLHPTITS*LFESWELNVFGLF 80
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 33.5 bits (75), Expect = 0.52
Identities = 21/71 (29%), Positives = 32/71 (44%)
Frame = +2
Query: 1029 GIEMRFASVEHPQSNGQVEAANKVILNGIKKRLGDAKGLWADELLTVVWAYNTTPQSTTG 1088
G + +S HP S+GQ EA NK ++ + W+ + YNT+ +
Sbjct: 65 GTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEYWYNTSYNISAA 244
Query: 1089 ETPFRLTYGVD 1099
TPF+ YG D
Sbjct: 245 MTPFKALYGRD 277
>AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 488
Score = 32.3 bits (72), Expect = 1.2
Identities = 14/37 (37%), Positives = 23/37 (61%)
Frame = +1
Query: 62 IKFLVVKCTSPYNVLIGRPSLNKLGAIISTRHLTVKY 98
I F V+ + Y+ L+GRP ++ GA+ ST H +K+
Sbjct: 1 ITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKF 111
>TC81098 homologue to GP|20373023|dbj|BAB91180. lipoic acid synthase
{Arabidopsis thaliana}, partial (44%)
Length = 842
Score = 30.0 bits (66), Expect = 5.7
Identities = 11/35 (31%), Positives = 19/35 (53%)
Frame = +3
Query: 919 CEKCQIYADLHRAPPEVLSSMSSAWPFAMWGVDIL 953
C C + ++ APP+ + ++A A WGVD +
Sbjct: 528 CRFCAVKTSINPAPPDPMEPENTAKAIASWGVDYI 632
>TC81434
Length = 363
Score = 24.3 bits (51), Expect(2) = 8.8
Identities = 11/24 (45%), Positives = 13/24 (53%)
Frame = +1
Query: 1200 AYHLEELSGRRIPRAWNAQHLRYY 1223
AY LE + R WNA HL+ Y
Sbjct: 154 AYKLEGRARNIYLRLWNATHLKLY 225
Score = 23.5 bits (49), Expect(2) = 8.8
Identities = 11/25 (44%), Positives = 15/25 (60%), Gaps = 2/25 (8%)
Frame = +2
Query: 1168 LVLRR--KAKGPDDSKLSPNWEGPY 1190
LVLR+ K KL+ +W+GPY
Sbjct: 53 LVLRKTEKVTNKKHGKLAASWKGPY 127
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 34,100,534
Number of Sequences: 36976
Number of extensions: 439099
Number of successful extensions: 1850
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 1835
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1844
length of query: 1224
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1117
effective length of database: 5,058,295
effective search space: 5650115515
effective search space used: 5650115515
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0306.1


