

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
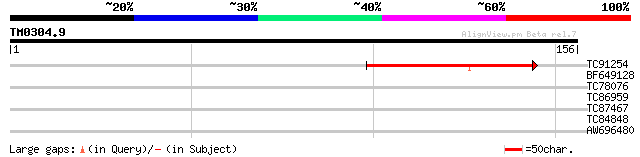
Query= TM0304.9
(156 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC91254 58 2e-09
BF649128 weakly similar to GP|17979475|gb AT3g25150/MJL12_9 {Ara... 27 4.4
TC78076 similar to GP|17017308|gb|AAL33611.1 SGT1a {Arabidopsis ... 26 5.7
TC86959 similar to GP|18086573|gb|AAL57710.1 AT5g15260/F8M21_150... 26 5.7
TC87467 similar to GP|17473624|gb|AAL38275.1 unknown protein {Ar... 26 5.7
TC84848 homologue to GP|21554112|gb|AAM63192.1 unknown {Arabidop... 25 9.8
AW696480 weakly similar to SP|P03211|EBN1_ EBNA-1 nuclear protei... 25 9.8
>TC91254
Length = 678
Score = 57.8 bits (138), Expect = 2e-09
Identities = 31/53 (58%), Positives = 35/53 (65%), Gaps = 6/53 (11%)
Frame = +2
Query: 99 AVRYLSYRLRATDGCRSDIIINLSVKL------LGSPDTCQTVIGIPVTAVRG 145
+VRYLSYRLR DG R IIN+SV+L + DTC TVIGIPVT RG
Sbjct: 20 SVRYLSYRLRGKDGSRGHAIINISVRLEITTLMSSNMDTCHTVIGIPVTTFRG 178
>BF649128 weakly similar to GP|17979475|gb AT3g25150/MJL12_9 {Arabidopsis
thaliana}, partial (25%)
Length = 527
Score = 26.6 bits (57), Expect = 4.4
Identities = 13/49 (26%), Positives = 26/49 (52%)
Frame = -2
Query: 82 CMIPALDIGFLNLRDSAAVRYLSYRLRATDGCRSDIIINLSVKLLGSPD 130
CMI LD F+N+ ++ +++ LR+ + S ++N+ + S D
Sbjct: 481 CMIRFLDFIFINISENIIQNKVAFLLRSKEETLSKFLLNVILSYHKSSD 335
>TC78076 similar to GP|17017308|gb|AAL33611.1 SGT1a {Arabidopsis thaliana},
partial (87%)
Length = 1493
Score = 26.2 bits (56), Expect = 5.7
Identities = 11/23 (47%), Positives = 16/23 (68%)
Frame = -1
Query: 19 SSYFITLTNLSNHLVYSYDGGGA 41
SS+F + +NLS LV+ +D G A
Sbjct: 998 SSFFTSASNLSQSLVFDFDDGYA 930
>TC86959 similar to GP|18086573|gb|AAL57710.1 AT5g15260/F8M21_150
{Arabidopsis thaliana}, partial (29%)
Length = 1380
Score = 26.2 bits (56), Expect = 5.7
Identities = 8/16 (50%), Positives = 13/16 (81%)
Frame = +2
Query: 37 DGGGAEEHMFRVPVDH 52
DGGG ++ +F++P DH
Sbjct: 890 DGGGIKKGLFKIPCDH 937
>TC87467 similar to GP|17473624|gb|AAL38275.1 unknown protein {Arabidopsis
thaliana}, partial (44%)
Length = 990
Score = 26.2 bits (56), Expect = 5.7
Identities = 10/23 (43%), Positives = 16/23 (69%)
Frame = -1
Query: 14 LGKYSSSYFITLTNLSNHLVYSY 36
+G S +YFITL+ L +H+ Y +
Sbjct: 513 IGNESFNYFITLSTLPSHVFYIF 445
>TC84848 homologue to GP|21554112|gb|AAM63192.1 unknown {Arabidopsis
thaliana}, partial (7%)
Length = 772
Score = 25.4 bits (54), Expect = 9.8
Identities = 9/22 (40%), Positives = 15/22 (67%)
Frame = +1
Query: 48 VPVDHTFFFDTQHSSLQLHLYN 69
+ V+HT +FD + L +HLY+
Sbjct: 112 ITVEHTNYFDQTPNFLNIHLYS 177
>AW696480 weakly similar to SP|P03211|EBN1_ EBNA-1 nuclear protein. [strain
B95-8 Human herpesvirus 4] {Epstein-barr virus},
partial (5%)
Length = 505
Score = 25.4 bits (54), Expect = 9.8
Identities = 18/53 (33%), Positives = 26/53 (48%)
Frame = +2
Query: 44 HMFRVPVDHTFFFDTQHSSLQLHLYNKRRILGPTQLGWCMIPALDIGFLNLRD 96
H FRV V + D Q LQLHL + R L W + +L + + +LR+
Sbjct: 131 HAFRVVVCNNRHLDLQQ*ELQLHLLSLR-------LTWHHLLSLRLTWHHLRE 268
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.138 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,884,438
Number of Sequences: 36976
Number of extensions: 68975
Number of successful extensions: 305
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 304
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 304
length of query: 156
length of database: 9,014,727
effective HSP length: 88
effective length of query: 68
effective length of database: 5,760,839
effective search space: 391737052
effective search space used: 391737052
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0304.9


