

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0304.11
(1797 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
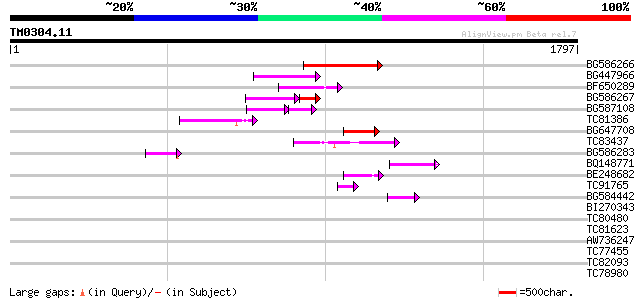
Score E
Sequences producing significant alignments: (bits) Value
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 201 2e-51
BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [impo... 163 6e-40
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 163 6e-40
BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At... 119 7e-40
BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - ... 98 7e-34
TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR re... 122 2e-27
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 105 1e-22
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 87 8e-17
BG586283 similar to GP|7267666|em RNA-directed DNA polymerase-li... 86 1e-16
BQ148771 82 1e-15
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 70 1e-11
TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative ret... 48 4e-05
BG584442 45 3e-04
BI270343 42 0.003
TC80480 similar to GP|4056493|gb|AAC98059.1| chloroplast lumen c... 37 0.053
TC81623 similar to PIR|T01257|T01257 probable GT-1-like transcri... 36 0.12
AW736247 weakly similar to PIR|T14619|T146 reverse transcriptase... 35 0.34
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 34 0.45
TC82093 34 0.45
TC78980 myb-related transcription factor [Medicago truncatula] 34 0.45
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 201 bits (511), Expect = 2e-51
Identities = 101/251 (40%), Positives = 154/251 (61%)
Frame = -3
Query: 931 NQYRPISCCNYLYKIISKVIVARLKGELNDIITPNQSAFIKDRLIQDNILIAHEIFHALQ 990
++YR I+ CN YKII+K++ R++ L II+P+QSAF+ R I DN+LI H+I H L+
Sbjct: 775 SEYRTIAPCNTQYKIIAKILSKRMQPLLRSIISPSQSAFVPGRAISDNVLITHKILHYLR 596
Query: 991 HRGREHKDHLAIKLDMSKAYDRVEWEFLEKALLAYGFCGPWVSLVMNLVRGVSYKYKVNG 1050
G + +A+K DM+KAYDR+ W FL + L GF G W+S +M V VSY + +NG
Sbjct: 595 QSGAKKHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFHGIWISWIMECVSTVSYSFLING 416
Query: 1051 YLSRKYIPQRGLRQGDPLSPYLFVLVMDVLSHMILQAKNQGHIQGIKLSPSTPCITHLFF 1110
+ +P RGLRQGDPLSPYLF+L +VLS + QA +G + G+K++ + P I HL F
Sbjct: 415 GPQGRVLPSRGLRQGDPLSPYLFILCTEVLSGLCQQALRKGTLPGVKVARNCPPINHLLF 236
Query: 1111 ADDSLVFAKATQQEAYHFVQLLNTFSSASSQRINTSKSSMSTSKHIPTLLKTQMTNIFQI 1170
ADD++ F K+ + +++ + +AS + IN +KS+++ S + ++ +I
Sbjct: 235 ADDTMFFGKSNASSCAILLSIMDKYRAASGRCIN*TKSAITFSSKTSQAIIDRVKGELKI 56
Query: 1171 PTWEEPGHYLG 1181
G YLG
Sbjct: 55 AKEGGTGKYLG 23
>BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 687
Score = 163 bits (412), Expect = 6e-40
Identities = 87/213 (40%), Positives = 123/213 (56%)
Frame = +2
Query: 773 WAQWGDRNTHFFHATTIQRRGRNRIHRLKDGTGSWIEGQEGVMENITDHFQLIFKSEQVT 832
W + GD+NT FFH+ QRR N I +LKD TG+W +G+E V + +F +F S T
Sbjct: 5 WLKDGDKNTKFFHSKASQRRKVNEIKKLKDETGNWCKGEENVERLLITYFNNLFTSSNPT 184
Query: 833 QIQECLSCVPKLIPTDLNNKLMETVTDEEIKVVAHSMDSLKAPGPDGLNGLFFKTHWNVV 892
I+E V + + + T+EE+ + M +KAPGPDGL LFF+ +W++V
Sbjct: 185 AIEETCEVVKGKLSHEHIVWCEKEFTEEEVLEAINQMHPVKAPGPDGLPALFFQKYWHIV 364
Query: 893 GPEVCSVVHSFFNTSTLSPCINETLVALVPKIPSLEMANQYRPISCCNYLYKIISKVIVA 952
G EV +V N S + +N+T + L+PK + YRPIS CN + KII+KVI
Sbjct: 365 GKEVQQMVLQVLNNSMETEELNKTFIVLIPKGKNPNTPKDYRPISLCNVVMKIITKVIAN 544
Query: 953 RLKGELNDIITPNQSAFIKDRLIQDNILIAHEI 985
R+K L D+I QSAF++ RLI DN LIA +
Sbjct: 545 RVKQTLPDVIDVEQSAFVQGRLITDNALIAWSV 643
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 163 bits (412), Expect = 6e-40
Identities = 81/201 (40%), Positives = 121/201 (59%)
Frame = +3
Query: 853 LMETVTDEEIKVVAHSMDSLKAPGPDGLNGLFFKTHWNVVGPEVCSVVHSFFNTSTLSPC 912
L T E+K SMDS KAPG DG N FFK WN++G V + FF T +
Sbjct: 12 LCSEFTAVEVKNALFSMDSSKAPGIDGYNVHFFKCSWNIIGDSVIDAILDFFKTGFMPKI 191
Query: 913 INETLVALVPKIPSLEMANQYRPISCCNYLYKIISKVIVARLKGELNDIITPNQSAFIKD 972
IN T V L+PK ++ +RPI+CC+ +YKIISK++ +R++G LN +++ NQSAF+K
Sbjct: 192 INCTYVTLLPKEVNVTSVKNFRPIACCSVIYKIISKILTSRMQGVLNSVVSENQSAFVKG 371
Query: 973 RLIQDNILIAHEIFHALQHRGREHKDHLAIKLDMSKAYDRVEWEFLEKALLAYGFCGPWV 1032
R+I DNI+++HE+ + +G + +K+D+ KAYD EW F++ +L GF +V
Sbjct: 372 RVIFDNIILSHELVKSYSRKGISPR--CMVKIDLXKAYDSXEWPFIKHLMLELGFPYKFV 545
Query: 1033 SLVMNLVRGVSYKYKVNGYLS 1053
+ VM + SY + NG L+
Sbjct: 546 NWVMAXLTTASYTFNXNGDLT 608
>BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (16%)
Length = 794
Score = 119 bits (299), Expect(2) = 7e-40
Identities = 58/171 (33%), Positives = 95/171 (54%)
Frame = +3
Query: 746 EMAQFKAQIRQLWRQEELFWSTRSRLQWAQWGDRNTHFFHATTIQRRGRNRIHRLKDGTG 805
E++ ++++ + + EE+FW +SRL W + GDRNT FFHA T RR +NRI L D
Sbjct: 78 ELSLLRSELNEEYHNEEIFWMQKSRLNWLRSGDRNTKFFHAVTKNRRAQNRILSLIDDDD 257
Query: 806 SWIEGQEGVMENITDHFQLIFKSEQVTQIQECLSCVPKLIPTDLNNKLMETVTDEEIKVV 865
+E + HF+L++ SE V E + +P ++ + N +LM ++ EE++
Sbjct: 258 KEWFVEEDLGRLADSHFKLLYSSEDVGITLEDWNSIPAIVTEEQNAQLMAQISREEVREA 437
Query: 866 AHSMDSLKAPGPDGLNGLFFKTHWNVVGPEVCSVVHSFFNTSTLSPCINET 916
++ K PGPDG+N FF+ W+ +G ++ S+ F T L IN+T
Sbjct: 438 VFDINPHKCPGPDGMNVFFFQQFWDTMGDDLTSMAQEFLRTGKLEEGINKT 590
Score = 64.7 bits (156), Expect(2) = 7e-40
Identities = 35/64 (54%), Positives = 43/64 (66%)
Frame = +1
Query: 920 LVPKIPSLEMANQYRPISCCNYLYKIISKVIVARLKGELNDIITPNQSAFIKDRLIQDNI 979
LVPK + ++RPIS CN YKI+SKV+ RLK L IIT Q+AF + +LI DNI
Sbjct: 601 LVPKKLEAKRLVEFRPISLCNVAYKIVSKVLSKRLKSVLPWIITETQAAFGRRQLISDNI 780
Query: 980 LIAH 983
LIAH
Sbjct: 781 LIAH 792
>BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - Arabidopsis
thaliana, partial (17%)
Length = 677
Score = 98.2 bits (243), Expect(2) = 7e-34
Identities = 49/137 (35%), Positives = 75/137 (53%)
Frame = +2
Query: 751 KAQIRQLWRQEELFWSTRSRLQWAQWGDRNTHFFHATTIQRRGRNRIHRLKDGTGSWIEG 810
K ++++ ++ EE +W +SR W GD N F+HA T QR RNRI L D G+WI
Sbjct: 11 KEKLQEAYKDEEDYWQQKSRNMWHISGDLNKKFYHALTKQRHARNRIVGLYDYDGNWITE 190
Query: 811 QEGVMENITDHFQLIFKSEQVTQIQECLSCVPKLIPTDLNNKLMETVTDEEIKVVAHSMD 870
++GV + D+F+ +F+ T L + I +N +L+ T+EE+++ M
Sbjct: 191 EQGVEKVAVDYFEDLFQRTTPTGFDGFLDEITSSITPQMNQRLLRLATEEEVRLALFIMH 370
Query: 871 SLKAPGPDGLNGLFFKT 887
KAPGPDG+ L F T
Sbjct: 371 PEKAPGPDGMTTLLFST 421
Score = 66.2 bits (160), Expect(2) = 7e-34
Identities = 34/87 (39%), Positives = 51/87 (58%)
Frame = +3
Query: 884 FFKTHWNVVGPEVCSVVHSFFNTSTLSPCINETLVALVPKIPSLEMANQYRPISCCNYLY 943
FF+ W+++ ++ +V+SF + L +N T + L+PK + RPIS CN Y
Sbjct: 411 FFQHSWHIIKMDLLKMVNSFLASGKLDTRLNTTNICLIPKKKRPTRMTELRPISLCNVGY 590
Query: 944 KIISKVIVARLKGELNDIITPNQSAFI 970
KIISKV+ RLK L +I+ QSAF+
Sbjct: 591 KIISKVLCQRLKVCLPSLISETQSAFV 671
>TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR retroelement
reverse transcriptase {Oryza sativa (japonica
cultivar-group)}, partial (1%)
Length = 798
Score = 122 bits (305), Expect = 2e-27
Identities = 74/257 (28%), Positives = 123/257 (47%), Gaps = 8/257 (3%)
Frame = -1
Query: 537 RRQALWDSISRLKPLAQGPWCCLGDFNETLYQHEKDGLHPQSQQSLNRFRSFVDSNALAN 596
+R+ LW +IS ++ + WCC+GDFN L HE G H ++ + F+ + D N L +
Sbjct: 798 KRRQLWSAISNIQTQHKLSWCCIGDFNTILGSHEHQGSHTPARLPMLDFQQWSDVNNLFH 619
Query: 597 IDIKGCKYTWLSNPREGFVTREKLDRVLSNWSWRSLYPHALVVALPVVSSDHSPLILDPV 656
+ +G +TW + R TR++LDR + N L + SDH P++ +
Sbjct: 618 LPTRGSAFTWTNGRRGRNNTRKRLDRSIVNQLMIDNCESLSACTLTKLRSDHFPILFELQ 439
Query: 657 PKH-RSGRQFKYEAFWNEHKDCSKVVEDAWNTTPQGENDWETLQNKVKSCQIGLTKWQKV 715
++ + FK+ W+ H DC +++ W G L K+K+ + L W K
Sbjct: 438 TQNIQFSSSFKFMKMWSAHPDCINIIKQCWANQVVG-CPMFVLNQKLKNLKEVLKVWNKN 262
Query: 716 TF-------SRADKEIEKLKARLSSMHNEDHHKIDWNEMAQFKAQIRQLWRQEELFWSTR 768
TF A KE++ ++ ++ S+ D +D + AQ + EE+FW +
Sbjct: 261 TFGNVHSQVDNAYKELDDIQVKIDSIGYSD-VLMDQEKAAQL--NLESALNIEEVFWHEK 91
Query: 769 SRLQWAQWGDRNTHFFH 785
S++ W GDRNT FFH
Sbjct: 90 SKVNWHCEGDRNTAFFH 40
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 105 bits (263), Expect = 1e-22
Identities = 53/115 (46%), Positives = 79/115 (68%)
Frame = +1
Query: 1058 PQRGLRQGDPLSPYLFVLVMDVLSHMILQAKNQGHIQGIKLSPSTPCITHLFFADDSLVF 1117
P++GLRQGDPLSPYLF+L +VLS ++ + N+ ++ GI+++ S P ITHL FADDSL+F
Sbjct: 4 PEKGLRQGDPLSPYLFILCANVLSGLLKREGNKQNLHGIQVARSDPKITHLLFADDSLLF 183
Query: 1118 AKATQQEAYHFVQLLNTFSSASSQRINTSKSSMSTSKHIPTLLKTQMTNIFQIPT 1172
A+A EA +Q+L+++ SAS Q +N KS +S S+++P K + I T
Sbjct: 184 ARANLTEAATIMQVLHSYQSASGQLVNFEKSEVSYSQNVPNQEKEMICQQIAIKT 348
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 86.7 bits (213), Expect = 8e-17
Identities = 88/348 (25%), Positives = 144/348 (41%), Gaps = 13/348 (3%)
Frame = +2
Query: 900 VHSFFNTSTLSPCINETLVALVPKIPSLEMANQYRPISCCNYLYKIISKVIVARLKGELN 959
V F L IN T +AL+PK+ + + N +RPIS LYKI+ K++ RL+ +
Sbjct: 11 VSEFHRNRKLFKGINSTFIALIPKVDNPQRLNDFRPISLVGSLYKILGKLLANRLRVVIG 190
Query: 960 DIITPNQSAFIKDRLIQDNILIAHEIFHALQHRGREHKDHLAIKLDMSKAYDRVEWEFLE 1019
+I+ QSAF+K+R I + + + L R R + K + + W
Sbjct: 191 SVISDAQSAFVKNRQILEMVFL*Q---MRLWMRLRN*R----------KIFCCLRWILKR 331
Query: 1020 KALLAYG-----------FCGPWVSLVMNLVRGVSYKYKVNGYLSRKYIPQRGLRQGDPL 1068
L+ G F W + V + VNG S + + L Q
Sbjct: 332 LITLSIGLIWILF*VGMSFLVLWRKWIKECVSTATTSVLVNG--SPTNVLMKSLVQTQLF 505
Query: 1069 SPYLFVLVMDVLSHMILQAKNQGHIQGIKLSPSTPCITHLFFADDSLVFAKATQQEAYHF 1128
+ Y F +V V+ ++HL FA+D+L+
Sbjct: 506 TRYSFGVVNPVV------------------------VSHLQFANDTLLLETKNWANIRAL 613
Query: 1129 VQLLNTFSSASSQRINTSKSSMSTSKHIPTLLKTQMTNIFQIPTWEEPGHYLGLPAVWGR 1188
L F + S ++N KS + P+ L ++ ++ + P YLG+P + G
Sbjct: 614 RAALVIF*AMSGLKVNFHKSGLVCVNIAPSWL-SEAASVLSWKVGKVPFLYLGMP-IEGN 787
Query: 1189 SKTRALAW--IKERMKEKIAGWKEVLLNQAGKEVLIKSVIQSIPTYAM 1234
S+ R W I R+K ++ GW L+ G+ VL+KSV+ S+ YA+
Sbjct: 788 SR-RLSFWEPIVNRIKARLTGWNSRFLSFGGRLVLLKSVLTSLSVYAL 928
>BG586283 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (7%)
Length = 774
Score = 85.9 bits (211), Expect = 1e-16
Identities = 43/124 (34%), Positives = 61/124 (48%), Gaps = 10/124 (8%)
Frame = +2
Query: 429 WNCRGVGASTTARELKEINSKQKPSILFLMETRARRNKIERLKFALHFSNFFCVDPIGTA 488
WNC+G+G+ T R L+E +K P I+FLMET+ + + ++ F+N F V P+G +
Sbjct: 218 WNCQGIGSDLTVRRLREFQTKDSPDIMFLMETKRKDEDVYKMYRGTEFTNHFTVPPVGLS 397
Query: 489 GGLALFWNKECTVEILDSNQNYIHVMITEKEKVIGAEYTF----------IYGEPLRQRR 538
GGLAL W VEIL S+ N I + +K Y + I G L R
Sbjct: 398 GGLALSWKDNVQVEILASSANVIDTKVNYNDKSFFVSYIYWCSPT*A*S*ILGRTLHHRG 577
Query: 539 QALW 542
W
Sbjct: 578 AKRW 589
Score = 35.8 bits (81), Expect = 0.15
Identities = 16/43 (37%), Positives = 20/43 (46%)
Frame = +3
Query: 531 GEPLRQRRQALWDSISRLKPLAQGPWCCLGDFNETLYQHEKDG 573
G P + R WD +S + G W GDFN+ L EK G
Sbjct: 519 GVPQPEHRAKFWDELSTIGAQRDGAWLLTGDFNDLLDNSEKVG 647
>BQ148771
Length = 680
Score = 82.4 bits (202), Expect = 1e-15
Identities = 50/165 (30%), Positives = 80/165 (48%), Gaps = 7/165 (4%)
Frame = -3
Query: 1205 IAGWKEVLLNQAGKEVLIKSVIQSIPTYAMAVTPFPKGFCKELDSLVARFWWSNNKKSRG 1264
+A WK L+ A + L KSVI+++P Y M T PK +E+ L +F W + + SR
Sbjct: 573 LANWKANHLSLARRVTLAKSVIEAVPLYPMMTTIIPKACIEEIQKLQRKFVWGDTEVSRR 394
Query: 1265 IHLKDWHSLTCSKKEGGMGFREFHLQNLAHMTKQAWRIHNTPHTLVAQVMKALYFPDSSF 1324
H W +++ K G+G R + N A + K W I++ ++L +VM+ Y S
Sbjct: 393 YHAVGWETMSKPKTIYGLGLRRLDVMNKACIMKLGWSIYSGSNSLCTEVMRGKYQRSESL 214
Query: 1325 LDA-TPKPGDSWMWRA------NQRQELLDRQRNLGCRGWSYHKL 1362
+ KP DS +W+A + L+D N W++ KL
Sbjct: 213 EEIFLEKPTDSSLWKALVKLWPEIERNLVDSNGN-----WNWEKL 94
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 69.7 bits (169), Expect = 1e-11
Identities = 42/125 (33%), Positives = 62/125 (49%)
Frame = +3
Query: 1059 QRGLRQGDPLSPYLFVLVMDVLSHMILQAKNQGHIQGIKLSPSTPCITHLFFADDSLVFA 1118
QRGL+QGDPL+P+LF+LV + +S ++ A N+ QG + ++HL +ADD+L
Sbjct: 21 QRGLKQGDPLAPFLFLLVAEGISGLMKNAVNRNLFQGFDVKRGGTRVSHLQYADDTLCIG 200
Query: 1119 KATQQEAYHFVQLLNTFSSASSQRINTSKSSMSTSKHIPTLLKTQMTNIFQIPTWEEPGH 1178
T + LL F AS ++N KSS+ ++P P
Sbjct: 201 MPTVDNLWTLKALLQGFEMASGLKVNFHKSSL-IGINVPRDFMEAACRFLNCREESIPFI 377
Query: 1179 YLGLP 1183
YLGLP
Sbjct: 378 YLGLP 392
>TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative retroelement
{Oryza sativa (japonica cultivar-group)}, partial (1%)
Length = 625
Score = 47.8 bits (112), Expect = 4e-05
Identities = 25/68 (36%), Positives = 36/68 (52%)
Frame = +2
Query: 1039 VRGVSYKYKVNGYLSRKYIPQRGLRQGDPLSPYLFVLVMDVLSHMILQAKNQGHIQGIKL 1098
V Y VN IP RGL+QGD LSPY+F++ ++ LS +I AK +G G +
Sbjct: 107 VESNDYYVLVNNDAVDPIIPSRGLQQGDHLSPYIFIICVEGLSFLIPHAKERGDTHGTSI 286
Query: 1099 SPSTPCIT 1106
P ++
Sbjct: 287 *RGAPPVS 310
Score = 41.6 bits (96), Expect = 0.003
Identities = 27/97 (27%), Positives = 46/97 (46%)
Frame = +3
Query: 1108 LFFADDSLVFAKATQQEAYHFVQLLNTFSSASSQRINTSKSSMSTSKHIPTLLKTQMTNI 1167
L F L + + A +L + S + I+ KS + S+++P +LKT +T I
Sbjct: 279 LLFEGAPLRSLRVEEHHAQIMKNILILYEEDSGKAISLRKS*IYCSRNVPDILKTSITYI 458
Query: 1168 FQIPTWEEPGHYLGLPAVWGRSKTRALAWIKERMKEK 1204
+ YLGLP++ GR +T + IK + +K
Sbjct: 459 LGVQFMLGTCKYLGLPSMIGRDRTTTFSSIKGGVWQK 569
>BG584442
Length = 775
Score = 45.1 bits (105), Expect = 3e-04
Identities = 29/102 (28%), Positives = 51/102 (49%), Gaps = 1/102 (0%)
Frame = +1
Query: 1198 KERMKEKIAGWKEVLLNQAGKEVLIKSVIQSIPTYAMAVTPFPKGFCKELDSLVARFWWS 1257
+ + +KI + L++ EV+IK +QSI +Y M++ E++ ++ F W
Sbjct: 364 RTKFDKKINF*RNKCLSKVM*EVMIKYALQSISSYVMSIFLLLNSQVDEIEKIMNTFSWV 543
Query: 1258 NNKKSR-GIHLKDWHSLTCSKKEGGMGFREFHLQNLAHMTKQ 1298
+ ++R G+H L K GGMGF +F N+ + KQ
Sbjct: 544 HVGENRKGMHWMS*EKLFVHKNYGGMGFTDFTTFNIPMLGKQ 669
>BI270343
Length = 619
Score = 41.6 bits (96), Expect = 0.003
Identities = 28/86 (32%), Positives = 39/86 (44%)
Frame = +1
Query: 385 EEKRISRCNGGGGIPQRTFLSKFCYHGWGGGH*SAPNVSMRLISWNCRGVGASTTARELK 444
+ K+I+ C G + F F W G A NV ISWNCRG+G +K
Sbjct: 379 QHKKINPCANGSFSAEELF---FIGRDWLPGLLGAMNV----ISWNCRGLGNVKAVPCIK 537
Query: 445 EINSKQKPSILFLMETRARRNKIERL 470
++ K ++ L+ET NKI L
Sbjct: 538 DLIRVYKSDVVILIETLVESNKISDL 615
>TC80480 similar to GP|4056493|gb|AAC98059.1| chloroplast lumen common
protein family {Arabidopsis thaliana}, partial (39%)
Length = 1295
Score = 37.4 bits (85), Expect = 0.053
Identities = 21/48 (43%), Positives = 28/48 (57%), Gaps = 1/48 (2%)
Frame = +3
Query: 281 KSRGPTPFYPTSCFELIP-KTSYSHHDPTPLEEPPQNHPKKPNPIKLL 327
+S P F P S F+L P K+S S +DP + PQN+P PNP + L
Sbjct: 90 RSHPPPQFLPPSSFKLPPIKSSSSQNDPA--FQKPQNNPTPPNPFQTL 227
>TC81623 similar to PIR|T01257|T01257 probable GT-1-like transcription
factor [imported] - Arabidopsis thaliana, partial (47%)
Length = 847
Score = 36.2 bits (82), Expect = 0.12
Identities = 17/35 (48%), Positives = 22/35 (62%)
Frame = +1
Query: 363 SEEGEEPTNH*GEGQHRGNCKKEEKRISRCNGGGG 397
SE+ EE N EG H+GN KK++K+ GGGG
Sbjct: 622 SEDEEELGNEESEGDHKGNIKKKKKKGKMIIGGGG 726
>AW736247 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (4%)
Length = 305
Score = 34.7 bits (78), Expect = 0.34
Identities = 17/54 (31%), Positives = 27/54 (49%), Gaps = 2/54 (3%)
Frame = +1
Query: 858 TDEEIKVVAHSMDSLKA--PGPDGLNGLFFKTHWNVVGPEVCSVVHSFFNTSTL 909
++EE++ DS + PGPDG+N F K +W ++ + VV F L
Sbjct: 124 SEEEVRKAVWDCDSSHS*NPGPDGVNFTFVKEYWELIKVDFLRVVMEFHTNGKL 285
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 34.3 bits (77), Expect = 0.45
Identities = 17/51 (33%), Positives = 28/51 (54%)
Frame = -3
Query: 1270 WHSLTCSKKEGGMGFREFHLQNLAHMTKQAWRIHNTPHTLVAQVMKALYFP 1320
WH L K GG+G R+ L N++ + K WR+ +L +V++ +Y P
Sbjct: 975 WHCLPRCK--GGLGVRDIRLVNVSLLAKWWWRLLQDQSSLWKEVLEDIYGP 829
>TC82093
Length = 569
Score = 34.3 bits (77), Expect = 0.45
Identities = 14/39 (35%), Positives = 22/39 (55%)
Frame = -1
Query: 756 QLWRQEELFWSTRSRLQWAQWGDRNTHFFHATTIQRRGR 794
+L Q E+ W +S+ W ++G RNT FFH + R +
Sbjct: 479 KLLSQNEMLWY*KSQ*NWVKYGSRNTWFFHNEDVN*RNQ 363
>TC78980 myb-related transcription factor [Medicago truncatula]
Length = 1526
Score = 34.3 bits (77), Expect = 0.45
Identities = 44/229 (19%), Positives = 93/229 (40%), Gaps = 10/229 (4%)
Frame = +2
Query: 562 FNETLYQHEKDGLHPQ--SQQSLNRFRSFVDSNALANIDIK----GCKYTWLSNPREGFV 615
FN++ Y+H + + ++ F +++ + D + G +WLSN
Sbjct: 557 FNDSKYEHILESFAEKLVKERPSPSFVMAASNSSYLHTDAQAPTPGLLPSWLSNSNNAAP 736
Query: 616 TREKLDRVLSNWSWRSLYPHALVVA----LPVVSSDHSPLILDPVPKHRSGRQFKYEAFW 671
R V SL P + PV D++PL+L V H + +
Sbjct: 737 VRPNSPSVTL-----SLSPSTVAAPPPWMQPVRGPDNAPLVLGNVAPHGAVLSYGESMVM 901
Query: 672 NEHKDCSKVVEDAWNTTPQGENDWETLQNKVKSCQIGLTKWQKVTFSRADKEIEKLKARL 731
+E DC K +E+ + + + ++V+ + + SR +++E+++A++
Sbjct: 902 SELVDCCKELEEVHHALAAHKKEAAWRLSRVE------LQLESEKASRRREKMEEIEAKI 1063
Query: 732 SSMHNEDHHKIDWNEMAQFKAQIRQLWRQEELFWSTRSRLQWAQWGDRN 780
++ E +D E +++ Q+ L R E T+ + QW ++
Sbjct: 1064KALREEQAVALDRIE-GEYREQLAGLRRDAE----TKEQKLTEQWAAKH 1195
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.331 0.143 0.482
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 65,550,364
Number of Sequences: 36976
Number of extensions: 1080360
Number of successful extensions: 6933
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 5014
Number of HSP's successfully gapped in prelim test: 240
Number of HSP's that attempted gapping in prelim test: 1609
Number of HSP's gapped (non-prelim): 5650
length of query: 1797
length of database: 9,014,727
effective HSP length: 110
effective length of query: 1687
effective length of database: 4,947,367
effective search space: 8346208129
effective search space used: 8346208129
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0304.11


