

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
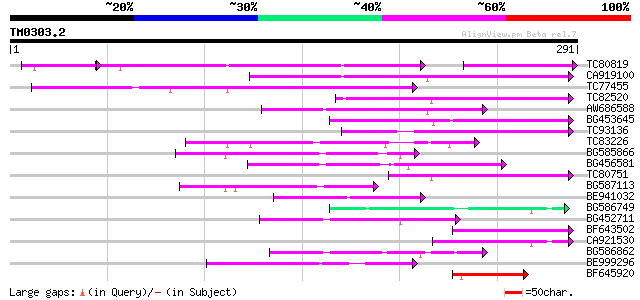
Query= TM0303.2
(291 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non... 74 1e-24
CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [i... 102 1e-22
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 94 5e-20
TC82520 73 1e-13
AW686588 66 1e-11
BG453645 62 3e-10
TC93136 55 2e-08
TC83226 weakly similar to PIR|G86419|G86419 probable reverse tra... 55 3e-08
BG585866 54 7e-08
BG456581 54 9e-08
TC80751 weakly similar to GP|13161403|dbj|BAB33036. CPRD49 {Vign... 50 8e-07
BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At... 49 2e-06
BE941032 49 3e-06
BG586749 weakly similar to GP|18138053|emb tropinone reductase I... 48 4e-06
BG452711 47 9e-06
BF643502 47 1e-05
CA921530 46 2e-05
BG586862 45 3e-05
BE999296 44 1e-04
BF645920 43 2e-04
>TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non-LTR
retroelement reverse transcriptase {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 1262
Score = 73.6 bits (179), Expect(3) = 1e-24
Identities = 53/175 (30%), Positives = 74/175 (42%), Gaps = 6/175 (3%)
Frame = +2
Query: 45 GSWEHGRWHWK-----FLWRRPLVGRELEWEKGLLDALGTVQLTKGVPDAWVWMPSNVGI 99
G + GR W + W R L E E + L L V D W W+ V
Sbjct: 374 GGADMGRLGWTVDGRAWEWTRRLFVWEEECVRECCILLNNFVLQDNVNDKWRWLLDPVNG 553
Query: 100 YSVNSAYHFLQEPTLPDTDEALVS-IWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVIT 158
YSV Y ++ T +D +LV +WH PS V F WR+L +R +++NL+ R V+
Sbjct: 554 YSVKVFYRYITS-TGHISDRSLVDDVWHKHIPSKVSLFVWRLLRNRLPTKDNLVHRGVLL 730
Query: 159 SLADAQCALCNDYVETGFHLLFSCTFSLSVWQGCLHWLGCSFIQPSSARDHLSHF 213
+ A C D E+ HL C S+W +WLG + R H F
Sbjct: 731 ATNAACVCGCVD-SESTTHLFLHCNVFCSLWSLVRNWLGIPSMSSGELRTHFIQF 892
Score = 49.7 bits (117), Expect(3) = 1e-24
Identities = 20/58 (34%), Positives = 33/58 (56%)
Frame = +1
Query: 234 WSVWLARNEVVFRGGAANIYSVLELVKRRSWQWNKANMKGFSYSSSDWFSNPLLCILG 291
W +W RN +F+ + +++ +K S+ W K+ GF+YS DW+ N LLC+ G
Sbjct: 946 WVIWKERNNRLFQNTVTAPFVLIDRIKLHSFLWLKSKQVGFAYSYLDWWKNSLLCMGG 1119
Score = 27.3 bits (59), Expect(3) = 1e-24
Identities = 18/44 (40%), Positives = 22/44 (49%), Gaps = 3/44 (6%)
Frame = +3
Query: 7 GDVTD--FWHDDWLGTGCLRGAFYRLYNLSRQKWNCVN-TLGSW 47
GD D FW+D W G LR + RL +L+ K V T G W
Sbjct: 267 GDGRDAFFWYDTWAGDVPLRLKYPRLLDLAMDKECKVGLTWGGW 398
>CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [imported] -
Sulfolobus solfataricus, partial (3%)
Length = 789
Score = 102 bits (255), Expect = 1e-22
Identities = 57/168 (33%), Positives = 80/168 (46%), Gaps = 2/168 (1%)
Frame = -2
Query: 124 IWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCT 183
IWH P V FAWR+L DR ++ NL+ R VI + A + C +E+ HL SC+
Sbjct: 584 IWHRQVPLKVSVFAWRLLRDRLPTKSNLIYRGVIPTEAGLCVSGCGA-LESAQHLFLSCS 408
Query: 184 FSLSVWQGCLHWLGCSFIQPSSARDHLSHF--RFGINKLQQRLALSIWLAVIWSVWLARN 241
+ S+W W+G + + DH F G NK Q IWL W +W RN
Sbjct: 407 YFASLWSLVRDWIGFVGVDTNVLSDHFVQFVHSTGGNKASQSFLQLIWLLCAWVLWTERN 228
Query: 242 EVVFRGGAANIYSVLELVKRRSWQWNKANMKGFSYSSSDWFSNPLLCI 289
+ F + +L+ VK S W KA F + + W+SNPL C+
Sbjct: 227 NMCFNDSITPLPRLLDKVKYLSLGWLKARNASFLFGTFSWWSNPLQCL 84
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 94.4 bits (233), Expect = 5e-20
Identities = 65/207 (31%), Positives = 92/207 (44%), Gaps = 9/207 (4%)
Frame = -2
Query: 12 FWHDDWLGTGCLRGAFYRLYNLSRQKWNCVNTLGSWEHGRWHWKFLWRRPLVGRELEWEK 71
FW D W + LR +F R + R L W+ WRR + EWEK
Sbjct: 676 FWKDAWDSSVPLRESFPRAFFPYRLLKMGCGDLWDMNAEGVRWRLYWRRLEL---FEWEK 506
Query: 72 G-LLDALGTVQ--LTKGVPDAWVWMPSNVGIYSVNSAYHFLQ-----EPTLPDTDEALV- 122
LL+ LG ++ + + D WVW P G++SVNS Y LQ E L +E +
Sbjct: 505 ERLLELLGRLEGVVLRYWADIWVWKPDKEGVFSVNSCYFLLQNLRLLEDRLSYEEEVIFR 326
Query: 123 SIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSC 182
+W S AP+ V F+W + LDR + NL KR+++ +C C ET HL C
Sbjct: 325 ELWKSKAPAKVLAFSWTLFLDRIPTMVNLGKRRLLRVEDSKRCVFCGCQDETVVHLFLHC 146
Query: 183 TFSLSVWQGCLHWLGCSFIQPSSARDH 209
V + + WL + I P + H
Sbjct: 145 DVISKV*REVMRWLNFNLISPPNLLIH 65
>TC82520
Length = 833
Score = 72.8 bits (177), Expect = 1e-13
Identities = 39/124 (31%), Positives = 56/124 (44%), Gaps = 2/124 (1%)
Frame = +2
Query: 168 CNDYVETGFHLLFSCTFSLSVWQGCLHWLGCSFIQPSSARDHLSHFRF--GINKLQQRLA 225
C D ET HL C S+W L WL + P+ R F G +
Sbjct: 317 CGDS-ETATHLFLHCDIFGSLWSHVLRWLHLLLVLPADIRQFFIQFTSMAGSPRFTHSFL 493
Query: 226 LSIWLAVIWSVWLARNEVVFRGGAANIYSVLELVKRRSWQWNKANMKGFSYSSSDWFSNP 285
+W A +W +W RN VF+ ++ + +E VK S+ W K FS+S DW+ +P
Sbjct: 494 QIMWFASVWVLWKKRNNRVFQNSLSDPSTFVEQVKMHSFLWLKFQQATFSFSYHDWWKHP 673
Query: 286 LLCI 289
LLC+
Sbjct: 674 LLCM 685
Score = 34.7 bits (78), Expect = 0.044
Identities = 23/97 (23%), Positives = 41/97 (41%), Gaps = 2/97 (2%)
Frame = +3
Query: 106 YHFLQEPTLPDTDEALVSIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQC 165
Y FL P + +W PS V F WR+ +R ++ NL++R V+ A
Sbjct: 132 YRFLTISGQPLDRNQVDDVWQKNIPSKVSMFVWRLFHNRLPTKVNLMQRHVLQQTHTA-- 305
Query: 166 ALCNDYVETGFHLLFSCTFSLSVW--QGCLHWLGCSF 200
+ + H+ F L+++ C+ ++ C F
Sbjct: 306 CISGVAIRKRQHICFYIVIFLALFGLMFCVGYIFCWF 416
>AW686588
Length = 567
Score = 66.2 bits (160), Expect = 1e-11
Identities = 38/118 (32%), Positives = 54/118 (45%), Gaps = 2/118 (1%)
Frame = +1
Query: 130 PSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVW 189
P V AWR++ DR ++ NL++R+ + A A C + ET HL C +VW
Sbjct: 145 PLKVSILAWRLIRDRLPTKANLVRRRCLAVEA-AGCVVGCGIAETANHLFLHCATFGAVW 321
Query: 190 QGCLHWLGCSFIQPSSARDHLSHF--RFGINKLQQRLALSIWLAVIWSVWLARNEVVF 245
Q W+G S P DH F G + ++ IWL +W VW RN +F
Sbjct: 322 QHIRAWIGVSGADPHDLSDHFIQFITCTGHTRARRSFMQLIWLLCVWMVWNERNNRLF 495
>BG453645
Length = 622
Score = 62.0 bits (149), Expect = 3e-10
Identities = 37/127 (29%), Positives = 58/127 (45%), Gaps = 2/127 (1%)
Frame = +3
Query: 165 CALCNDYVETGFHLLFSCTFSLSVWQGCLHWLGCSFIQPSSARDHLSHFRFG--INKLQQ 222
C LCN VET H+ C + VW + W +F+ P + H + G N++ +
Sbjct: 33 CVLCNLKVETTTHIFLHCDVASLVWFRVMKWFDVNFLIPPNLFVHWECWSEGGSANRVTK 212
Query: 223 RLALSIWLAVIWSVWLARNEVVFRGGAANIYSVLELVKRRSWQWNKANMKGFSYSSSDWF 282
L +W IW +W RN+++F+G V+E +K S +W S +W
Sbjct: 213 GHWL-VWHTTIWVLWAKRNDLIFKGLNCVAEDVIEEIKVLS*RWMLERSSTPSCFFYEWS 389
Query: 283 SNPLLCI 289
NP LC+
Sbjct: 390 WNPRLCL 410
>TC93136
Length = 722
Score = 55.5 bits (132), Expect = 2e-08
Identities = 34/119 (28%), Positives = 51/119 (42%)
Frame = +1
Query: 171 YVETGFHLLFSCTFSLSVWQGCLHWLGCSFIQPSSARDHLSHFRFGINKLQQRLALSIWL 230
Y ET HL C F SVW L WL S + FR + + + IW
Sbjct: 256 YNETSSHLFLHCPFLSSVWSKILGWLYYS--------KFVCVFRLLERRSRTKGVWLIWH 411
Query: 231 AVIWSVWLARNEVVFRGGAANIYSVLELVKRRSWQWNKANMKGFSYSSSDWFSNPLLCI 289
A IW +W N +F+ + I ++E +K SW+W+ +K +W +P C+
Sbjct: 412 ATIWVIWKGINNRIFKNISKAIDEIVEEIKVLSWRWSLTRLKCPPCFYYEWCWDPGDCV 588
>TC83226 weakly similar to PIR|G86419|G86419 probable reverse transcriptase
100033-105622 [imported] - Arabidopsis thaliana, partial
(2%)
Length = 885
Score = 55.1 bits (131), Expect = 3e-08
Identities = 51/169 (30%), Positives = 75/169 (44%), Gaps = 18/169 (10%)
Frame = +3
Query: 91 VWMPSNVGIYSVNSAYHFLQ-------EPTLPDTDEALV--SIWHSFAPSNVKGFAWRVL 141
+WM + GIYSV S Y+ L+ T +DE L+ IW K WR+L
Sbjct: 3 MWMHNPTGIYSVKSGYNTLRTWQTQQINNTSTSSDETLIWKKIWSLHTIPRHKVLLWRIL 182
Query: 142 LDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVWQG---CLHWLGC 198
D R +L KR + C C+ ET HL SC S VW G C+++
Sbjct: 183 NDSLPVRSSLRKRGI---QCYPLCPRCHSKTETITHLFMSCPLSKRVWFGSNLCINF--- 344
Query: 199 SFIQPSSARDHLSHFRFGINKLQQRL-----ALSIWL-AVIWSVWLARN 241
D+L + F IN L + + ++I + A+I+++W ARN
Sbjct: 345 ---------DNLPNPNF-IN*LYEAIL*KDECITI*IAAIIYNLWHARN 461
>BG585866
Length = 828
Score = 53.9 bits (128), Expect = 7e-08
Identities = 36/130 (27%), Positives = 60/130 (45%), Gaps = 5/130 (3%)
Frame = +3
Query: 86 VPDAWVWMPSNVGIYSVNSAYHFL--QEPTLPDTDEALVSIWHSFAPSNVKGFAWRVLLD 143
+ DA++W ++ G+Y+ S Y ++ Q T+ + + IW P K F W +
Sbjct: 342 IGDAYIWPHNSNGVYTAKSGYSWILSQTETVNYNNSSWSWIWRLKIPEKYKFFLWLACHN 521
Query: 144 RFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVWQGCLHWLGCS---F 200
+ L R ++ S A C+ C ++ E+ FH + C FS +W H +G S F
Sbjct: 522 AVPTLSLLNHRNMVNS---AICSRCGEHEESFFHCVRDCRFSKIIW----HKIGFSSPDF 680
Query: 201 IQPSSARDHL 210
SS +D L
Sbjct: 681 FSSSSVQDWL 710
>BG456581
Length = 683
Score = 53.5 bits (127), Expect = 9e-08
Identities = 38/137 (27%), Positives = 64/137 (45%), Gaps = 4/137 (2%)
Frame = +2
Query: 123 SIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSC 182
+IW+S P + WR+ DR + +NL R + C+ C + VET HL C
Sbjct: 77 TIWNSCIPPSHSFICWRLAHDRLPTDDNLSSRGCALV---SMCSFCLEQVETSDHLFLRC 247
Query: 183 TFSLSVWQGCLHWLGCSFIQP----SSARDHLSHFRFGINKLQQRLALSIWLAVIWSVWL 238
F +++W WL CS ++ SS + LS + + L ++ + ++ S+W
Sbjct: 248 KFVVTLWS----WL-CSQLRVGLDFSSFKALLSSLPRHCSSQVRDLYVAAVVHMVHSIWW 412
Query: 239 ARNEVVFRGGAANIYSV 255
ARN V F + ++V
Sbjct: 413 ARNNVRFSSAKVSAHAV 463
>TC80751 weakly similar to GP|13161403|dbj|BAB33036. CPRD49 {Vigna
unguiculata}, partial (52%)
Length = 1156
Score = 50.4 bits (119), Expect = 8e-07
Identities = 28/97 (28%), Positives = 47/97 (47%), Gaps = 2/97 (2%)
Frame = +1
Query: 195 WLGCSFIQPSSARDHLSHFRF--GINKLQQRLALSIWLAVIWSVWLARNEVVFRGGAANI 252
+LG S P DHL G ++ + L +W + +W +W R++++F ++
Sbjct: 700 FLGVSSTDPPCIPDHLIQLVQ*GGYSRSIRSTLLLVWFSYMWVIWKERSKMIFNQKKDHV 879
Query: 253 YSVLELVKRRSWQWNKANMKGFSYSSSDWFSNPLLCI 289
+ +LE VK S+ W KA F + NPLLC+
Sbjct: 880 HQLLEKVKLLSF*WLKATNSTFPFDYHSRRLNPLLCM 990
>BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (10%)
Length = 767
Score = 49.3 bits (116), Expect = 2e-06
Identities = 30/112 (26%), Positives = 47/112 (41%), Gaps = 10/112 (8%)
Frame = -3
Query: 88 DAWVWMPSNVGIYSVNSAYHFL--------QEPTL--PDTDEALVSIWHSFAPSNVKGFA 137
D++ W S G YSV S Y+ Q T+ P D+ +W V+ F
Sbjct: 426 DSYSWEYSKSGHYSVKSGYYVQTNIIAAANQRGTVDQPSLDDLYQRVWKYNTSPKVRHFL 247
Query: 138 WRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVW 189
WR + + + N+ R + D C+ C ET H+LF C ++ +W
Sbjct: 246 WRCISNSLPTAANMRSRHISK---DGSCSRCGMESETVNHILFQCPYARLIW 100
>BE941032
Length = 435
Score = 48.5 bits (114), Expect = 3e-06
Identities = 25/78 (32%), Positives = 38/78 (48%)
Frame = +2
Query: 136 FAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVWQGCLHW 195
F W VLL+RF +++NLLKR VI+++ + C + + HL C F +W W
Sbjct: 152 FLWCVLLNRFPTKDNLLKRGVISAIYQSCVGECGNLYD-ATHLFLHCNFFRQIWINVSDW 328
Query: 196 LGCSFIQPSSARDHLSHF 213
L + D L+ F
Sbjct: 329 LSFVMVTLLRISDRLAQF 382
>BG586749 weakly similar to GP|18138053|emb tropinone reductase I {Calystegia
sepium}, partial (11%)
Length = 787
Score = 48.1 bits (113), Expect = 4e-06
Identities = 31/125 (24%), Positives = 50/125 (39%), Gaps = 2/125 (1%)
Frame = -2
Query: 165 CALCNDYVETGFHLLFSCTFSLSVWQGCLHWLGCSFIQPSSARDHLSHFRFGINKLQQRL 224
C LC E HL C + VW L WLG + P H + + R
Sbjct: 531 CVLCEGVPEVSEHLFLQCA-TAKVWADLLRWLGFDYHTPPDLFFHWKWWNEMSGNKKTRK 355
Query: 225 ALSIWLAVIWSVWLARNEVVFRGGAANIYSVLELVKRRSWQW--NKANMKGFSYSSSDWF 282
I +W RN+++F + + ++E +K +W+W N+ N+ + +W
Sbjct: 354 GY*I-------IWRVRNDLIFNNSRSGMDELVEAIKVLAWRWVLNRLNLPACMF--YEWI 202
Query: 283 SNPLL 287
N LL
Sbjct: 201 WNFLL 187
>BG452711
Length = 672
Score = 47.0 bits (110), Expect = 9e-06
Identities = 39/115 (33%), Positives = 50/115 (42%), Gaps = 12/115 (10%)
Frame = +3
Query: 129 APSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSV 188
APSN+K F ++ D R LL +I+S C +CN + H LFSCT + V
Sbjct: 309 APSNIKLFL*QL*RDYVHFRSILLFCNLISSNL---CPICNQRSQDMLHALFSCTRAKDV 479
Query: 189 WQGCLHWLGCS------------FIQPSSARDHLSHFRFGINKLQQRLALSIWLA 231
W CLH L S FI D LS RFG+ L +S L+
Sbjct: 480 WSFCLHELVTSSNQDNLITWMKEFISNIGPLDPLSCGRFGVLTTNVSLXISSXLS 644
>BF643502
Length = 631
Score = 46.6 bits (109), Expect = 1e-05
Identities = 23/62 (37%), Positives = 31/62 (49%)
Frame = -1
Query: 228 IWLAVIWSVWLARNEVVFRGGAANIYSVLELVKRRSWQWNKANMKGFSYSSSDWFSNPLL 287
IWL +W VW RN V+F I +L+ +K S W KA F + SNPL+
Sbjct: 490 IWLLCVWVVWNERNNVMFNHVVTPIPCLLDKIKLLSLGWLKAKNATFVFGIQQGRSNPLV 311
Query: 288 CI 289
C+
Sbjct: 310 CL 305
>CA921530
Length = 793
Score = 45.8 bits (107), Expect = 2e-05
Identities = 26/74 (35%), Positives = 36/74 (48%), Gaps = 2/74 (2%)
Frame = -1
Query: 218 NKLQQRLALSIWLAVIWSVWLARNEVVFRGGAANIYSVLELVKRRSWQW--NKANMKGFS 275
NK +R IW A IW +W A N +F A ++E VK SW+W N+ N+
Sbjct: 622 NKKIRRGLWLIWHATIWILWKAWNNRIFNNQAVEAEDIIEEVKVLSWRWTFNRINIPVCM 443
Query: 276 YSSSDWFSNPLLCI 289
Y +W NP C+
Sbjct: 442 Y--YEWCWNPKYCL 407
>BG586862
Length = 804
Score = 45.4 bits (106), Expect = 3e-05
Identities = 34/114 (29%), Positives = 55/114 (47%), Gaps = 2/114 (1%)
Frame = -1
Query: 134 KGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVWQGCL 193
K F WR+L + ++ L KR + SL C C +ET HL +C + W G
Sbjct: 624 KSFLWRLLHNALPVKDELHKRGIRCSLL---CPRCESKIETVQHLFLNCEVTQKEWFG-- 460
Query: 194 HWLGCSFIQPSSARDH--LSHFRFGINKLQQRLALSIWLAVIWSVWLARNEVVF 245
LG +F H +++F I K + +++ A+++S+W ARN+ VF
Sbjct: 459 SQLGINFHSSGVLHFHDWITNF---ILKNDEETIIAL-TALLYSIWHARNQKVF 310
>BE999296
Length = 384
Score = 43.5 bits (101), Expect = 1e-04
Identities = 31/108 (28%), Positives = 46/108 (41%)
Frame = -1
Query: 102 VNSAYHFLQEPTLPDTDEALVSIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLA 161
++ AY FL P E + S+WH+ P V F RVL + +++N ++R+VI
Sbjct: 306 IHGAYQFLMSADAPLDREYIDSVWHNHIPLKVCFFVLRVLRNCLPTKDNFVRRRVIHE-E 130
Query: 162 DAQCALCNDYVETGFHLLFSCTFSLSVWQGCLHWLGCSFIQPSSARDH 209
C + ET T L +W WL S + RDH
Sbjct: 129 HMLCPTGCSFKET--------TDDLFLWPLV*QWLHIS*VNSCVLRDH 10
>BF645920
Length = 631
Score = 42.7 bits (99), Expect = 2e-04
Identities = 16/42 (38%), Positives = 26/42 (61%), Gaps = 3/42 (7%)
Frame = -3
Query: 228 IWL---AVIWSVWLARNEVVFRGGAANIYSVLELVKRRSWQW 266
+WL +W++W ARN ++F G + S++E VK SW+W
Sbjct: 626 LWLISTTTVWAIWRARNNIIFNEGIVEVDSMVEEVKVLSWRW 501
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.138 0.483
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,538,681
Number of Sequences: 36976
Number of extensions: 250058
Number of successful extensions: 1955
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 1894
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1931
length of query: 291
length of database: 9,014,727
effective HSP length: 95
effective length of query: 196
effective length of database: 5,502,007
effective search space: 1078393372
effective search space used: 1078393372
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0303.2


