

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
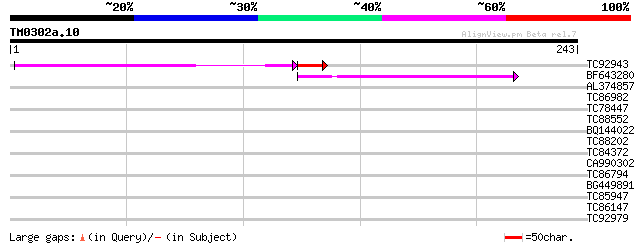
Query= TM0302a.10
(243 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC92943 107 6e-25
BF643280 42 2e-04
AL374857 39 0.002
TC86982 similar to GP|10946337|gb|AAG24863.1 CONSTANS-like prote... 30 0.85
TC78447 similar to GP|7768151|emb|CAB90633.1 protein phpsphatase... 29 1.4
TC88552 similar to PIR|T51260|T51260 hypothetical protein T8M16_... 29 1.9
BQ144022 homologue to GP|22831315|dbj hypothetical protein~predi... 28 3.2
TC88202 homologue to GP|21615421|emb|CAD33930. arabinogalactan p... 28 4.2
TC84372 similar to GP|15146181|gb|AAK83574.1 AT3g14070/MAG2_2 {A... 27 5.5
CA990302 weakly similar to GP|10177640|db contains similarity to... 27 7.2
TC86794 similar to PIR|T06499|T06499 Rieske [2Fe-2S] iron-sulfur... 27 7.2
BG449891 similar to GP|17901818|gb 32 kDa ferritin subunit {Gall... 27 7.2
TC85947 similar to PIR|S52261|S52261 NADH dehydrogenase (ubiquin... 27 9.4
TC86147 GP|9758956|dbj|BAB09343.1 histone H2A {Arabidopsis thali... 27 9.4
TC92979 similar to GP|20127014|gb|AAM10934.1 putative bHLH trans... 27 9.4
>TC92943
Length = 672
Score = 107 bits (267), Expect(2) = 6e-25
Identities = 59/121 (48%), Positives = 72/121 (58%)
Frame = +1
Query: 3 KAGRGKTTGTSVTKKKMKSATGKLPLCIPLDKMVAVGPGAPDFVTEISVLVKKKAPFNVK 62
K GRG+T G SV KKK KSATGKL IP DKMVAVG GA +FVTEIS++V K APFNVK
Sbjct: 310 KVGRGRTVGLSVAKKKKKSATGKLHDVIPTDKMVAVGSGAANFVTEISIVVLKNAPFNVK 489
Query: 63 GWKKVPDTAKDHILSQVFVNIII*FIFLFSFVLIQYLQYISCNCWVQDTFDIDQTEHNKE 122
W+K+P ++ I+S+V DTFDID T+HN++
Sbjct: 490 KWRKIPQETENKIVSKVL-----------------------------DTFDIDNTKHNRD 582
Query: 123 V 123
V
Sbjct: 583 V 585
Score = 23.5 bits (49), Expect(2) = 6e-25
Identities = 9/13 (69%), Positives = 12/13 (92%)
Frame = +2
Query: 124 ILNTAKRLYGSHR 136
I++TAKRLY +HR
Sbjct: 587 IIDTAKRLYRNHR 625
>BF643280
Length = 582
Score = 42.4 bits (98), Expect = 2e-04
Identities = 29/95 (30%), Positives = 46/95 (47%)
Frame = +2
Query: 124 ILNTAKRLYGSHRGKLHEHFKKYETNEMALEHKPDEISEEDWEHLVDKFSSPTYKEMSAR 183
I+N A R Y K HF KY+ L+++P+ I E ++ L+ +S +E+S +
Sbjct: 272 IINDAWRWYKWLIKK--NHFTKYKNLCERLKNRPNSIPEAHFKKLMTYWSYEAIREISHQ 445
Query: 184 NKANKSKQVINHRCGRKSFQAVSYDARDPETQKEP 218
N N KQ H+ G SF + R + +EP
Sbjct: 446 NAKNIDKQKWRHQAGPISFAVIRERLRATKDDREP 550
>AL374857
Length = 492
Score = 38.9 bits (89), Expect = 0.002
Identities = 22/91 (24%), Positives = 47/91 (51%), Gaps = 2/91 (2%)
Frame = +1
Query: 155 HKPDEISEEDWEHLVDKFSSPTYKEMSARNKANKSKQVIN--HRCGRKSFQAVSYDARDP 212
H P + E W LVD + P + + S N+ N++ + + H G SF+ +
Sbjct: 91 HGPKGLKPEVWNGLVDIWLKPEWTKKSDANRCNRAAKPDSALHTGGSISFRE-HKKRMEE 267
Query: 213 ETQKEPNYQDLWRMTHTNNNGEWVNEASREV 243
E ++E +Y+D++ H + +G +++ S+++
Sbjct: 268 EMKQEVSYRDVYAHVHKSKDGGYISRRSKKL 360
>TC86982 similar to GP|10946337|gb|AAG24863.1 CONSTANS-like protein {Ipomoea
nil}, partial (54%)
Length = 1643
Score = 30.0 bits (66), Expect = 0.85
Identities = 14/36 (38%), Positives = 20/36 (54%), Gaps = 2/36 (5%)
Frame = +1
Query: 89 FLFSFVLIQYLQYISCNCWVQD--TFDIDQTEHNKE 122
+LFS + +YL + CN W D TF + T H+ E
Sbjct: 637 YLFSGEVDEYLDLVDCNSWGGDENTFTTNNTHHHDE 744
>TC78447 similar to GP|7768151|emb|CAB90633.1 protein phpsphatase 2C (PP2C)
{Fagus sylvatica}, partial (37%)
Length = 833
Score = 29.3 bits (64), Expect = 1.4
Identities = 11/27 (40%), Positives = 22/27 (80%)
Frame = +2
Query: 76 LSQVFVNIII*FIFLFSFVLIQYLQYI 102
+ Q+ ++++I FIF FS +LIQ+L+++
Sbjct: 23 IKQIKISLLIIFIFQFS*ILIQWLKFV 103
>TC88552 similar to PIR|T51260|T51260 hypothetical protein T8M16_50 -
Arabidopsis thaliana, partial (25%)
Length = 1588
Score = 28.9 bits (63), Expect = 1.9
Identities = 22/101 (21%), Positives = 47/101 (45%), Gaps = 9/101 (8%)
Frame = +2
Query: 126 NTAKRLYG-SHRGKLHEHFKKYE--TNEMALEHKPDEISEE------DWEHLVDKFSSPT 176
++++ YG S K + ++K+E ++ A + + D++ EE D+ +DK+S
Sbjct: 377 HSSRTSYGHSRHDKYADEYRKHERLSSRSARDSRGDQMREESDSRSKDYGRSMDKYSREK 556
Query: 177 YKEMSARNKANKSKQVINHRCGRKSFQAVSYDARDPETQKE 217
Y+ R+K ++ + K + DARD + +
Sbjct: 557 YERSDFRSKDRDREKRATYEEVEKEKRTRDGDARDEKKDSQ 679
>BQ144022 homologue to GP|22831315|dbj hypothetical protein~predicted by
GlimmerM etc. {Oryza sativa (japonica cultivar-group)},
partial (16%)
Length = 1286
Score = 28.1 bits (61), Expect = 3.2
Identities = 15/44 (34%), Positives = 22/44 (49%)
Frame = +2
Query: 147 ETNEMALEHKPDEISEEDWEHLVDKFSSPTYKEMSARNKANKSK 190
ET + L H P S +H K S PT++ +ARNK + +
Sbjct: 47 ETTQRFLGHTPQNFSSLKRKH---KTSKPTHERRTARNKKQRGQ 169
>TC88202 homologue to GP|21615421|emb|CAD33930. arabinogalactan protein
{Cicer arietinum}, complete
Length = 628
Score = 27.7 bits (60), Expect = 4.2
Identities = 12/21 (57%), Positives = 16/21 (76%)
Frame = +2
Query: 85 I*FIFLFSFVLIQYLQYISCN 105
I*F + FSF+LI+Y Y+S N
Sbjct: 524 I*FFYYFSFMLIKYYFYLSIN 586
>TC84372 similar to GP|15146181|gb|AAK83574.1 AT3g14070/MAG2_2 {Arabidopsis
thaliana}, partial (35%)
Length = 755
Score = 27.3 bits (59), Expect = 5.5
Identities = 14/54 (25%), Positives = 25/54 (45%)
Frame = -1
Query: 30 IPLDKMVAVGPGAPDFVTEISVLVKKKAPFNVKGWKKVPDTAKDHILSQVFVNI 83
I D + +V + D + K APF ++ K +PD+A H++ N+
Sbjct: 662 IHXDAVTSVRKSSTDAAENVGT---KSAPFMIEVIKLLPDSANSHVIQAAGANV 510
>CA990302 weakly similar to GP|10177640|db contains similarity to
heparanase~gene_id:MGG23.2 {Arabidopsis thaliana},
partial (42%)
Length = 786
Score = 26.9 bits (58), Expect = 7.2
Identities = 9/14 (64%), Positives = 11/14 (78%)
Frame = +1
Query: 52 LVKKKAPFNVKGWK 65
+ KK+ PFN KGWK
Sbjct: 559 ITKKRVPFNSKGWK 600
>TC86794 similar to PIR|T06499|T06499 Rieske [2Fe-2S] iron-sulfur protein
tic55 - garden pea, partial (11%)
Length = 812
Score = 26.9 bits (58), Expect = 7.2
Identities = 16/74 (21%), Positives = 32/74 (42%), Gaps = 5/74 (6%)
Frame = -2
Query: 142 HFKKYETNEMALEHKPDEISEEDWEHLVDKFSSPTYKEMSARNKANKSKQVI-----NHR 196
HF K+ + + D IS ++EH+ S + + M + + ++++I R
Sbjct: 436 HFSKFGKYKPSSSRSSDGISPSEYEHMRTAISKMSAENMELKERLKTNEELIRASQEESR 257
Query: 197 CGRKSFQAVSYDAR 210
R+ Q D+R
Sbjct: 256 LAREQAQHSQEDSR 215
>BG449891 similar to GP|17901818|gb 32 kDa ferritin subunit {Galleria
mellonella}, partial (74%)
Length = 649
Score = 26.9 bits (58), Expect = 7.2
Identities = 18/78 (23%), Positives = 35/78 (44%), Gaps = 2/78 (2%)
Frame = +1
Query: 142 HFKKYETNEMALEHKPDEISEEDWEHLVDKFSSPTYK--EMSARNKANKSKQVINHRCGR 199
+F Y+TN + ++S+E WE +D T + EM+ ++ + N+
Sbjct: 358 YFNNYQTNRLGFSKLFKKLSDEAWEKTIDIIKHVTMRGDEMNFDQRSTMKVERKNYTVEL 537
Query: 200 KSFQAVSYDARDPETQKE 217
+A+ A+ +TQ E
Sbjct: 538 HELEAL---AKALDTQXE 582
>TC85947 similar to PIR|S52261|S52261 NADH dehydrogenase (ubiquinone) (EC
1.6.5.3) flavoprotein 1 precursor - potato, partial
(44%)
Length = 1086
Score = 26.6 bits (57), Expect = 9.4
Identities = 12/36 (33%), Positives = 20/36 (55%)
Frame = -2
Query: 9 TTGTSVTKKKMKSATGKLPLCIPLDKMVAVGPGAPD 44
+T T+ + + S +LPLCI + + +GP PD
Sbjct: 698 STVTTPWSRPLTSGLQQLPLCILFNPSLKLGPEMPD 591
>TC86147 GP|9758956|dbj|BAB09343.1 histone H2A {Arabidopsis thaliana},
partial (94%)
Length = 678
Score = 26.6 bits (57), Expect = 9.4
Identities = 19/56 (33%), Positives = 27/56 (47%), Gaps = 10/56 (17%)
Frame = +1
Query: 4 AGRGKTTGTSVTKKKMKSATGKLPLCIPLDKMV----------AVGPGAPDFVTEI 49
AGRGKT G S + KK S + K L P+ ++ VG GAP ++ +
Sbjct: 103 AGRGKTLG-SGSAKKATSRSSKAGLQFPVGRIARFLKAGKYAERVGAGAPVYLAAV 267
>TC92979 similar to GP|20127014|gb|AAM10934.1 putative bHLH transcription
factor {Arabidopsis thaliana}, partial (15%)
Length = 744
Score = 26.6 bits (57), Expect = 9.4
Identities = 9/22 (40%), Positives = 14/22 (62%)
Frame = +3
Query: 55 KKAPFNVKGWKKVPDTAKDHIL 76
+K + +K WK+ P AK H+L
Sbjct: 441 QKGHWKIKSWKQKPQKAKGHVL 506
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.133 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,831,861
Number of Sequences: 36976
Number of extensions: 113147
Number of successful extensions: 717
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 713
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 717
length of query: 243
length of database: 9,014,727
effective HSP length: 93
effective length of query: 150
effective length of database: 5,575,959
effective search space: 836393850
effective search space used: 836393850
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0302a.10


