

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
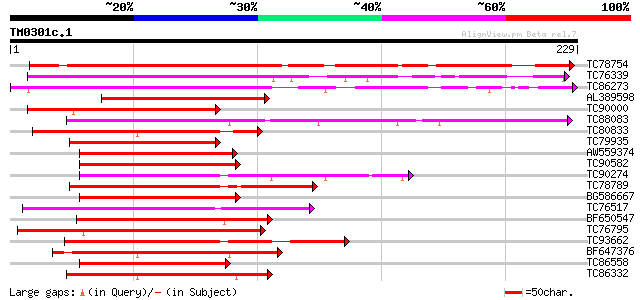
Query= TM0301c.1
(229 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78754 homologue to GP|4099921|gb|AAD09248.1| EREBP-3 homolog {... 274 2e-74
TC76339 homologue to GP|4099914|gb|AAD00708.1| ethylene-responsi... 171 2e-43
TC86273 similar to SP|O80340|ERF4_ARATH Ethylene responsive elem... 167 4e-42
AL389598 homologue to GP|4099914|gb|A ethylene-responsive elemen... 123 7e-29
TC90000 homologue to GP|9294548|dbj|BAB02811.1 ethylene responsi... 114 3e-26
TC88083 similar to GP|4587373|dbj|BAA76734.1 ethylene responsive... 111 3e-25
TC80833 weakly similar to PIR|D86175|D86175 hypothetical protein... 110 3e-25
TC79935 homologue to GP|15207792|dbj|BAB62912. ERF domain protei... 107 3e-24
AW559374 similar to GP|17381278|gb| AT4g34410/F10M10_180 {Arabid... 103 7e-23
TC90582 similar to GP|17381278|gb|AAL36057.1 AT4g34410/F10M10_18... 102 2e-22
TC90274 similar to GP|10177219|dbj|BAB10294. contains similarity... 101 2e-22
TC78789 homologue to PIR|T48580|T48580 hypothetical protein T31B... 100 5e-22
BG586667 similar to GP|17381278|gb| AT4g34410/F10M10_180 {Arabid... 100 5e-22
TC76517 similar to GP|11994632|dbj|BAB02769. AP2 domain transcri... 99 2e-21
BF650547 similar to GP|9758388|dbj| AP2 domain transcription fac... 98 2e-21
TC76795 similar to GP|18496063|emb|CAD21849. ethylene responsive... 97 4e-21
TC93662 similar to GP|10177219|dbj|BAB10294. contains similarity... 96 9e-21
BF647376 similar to GP|7939533|dbj| Nicotiana EREBP-3-like prote... 96 1e-20
TC86558 similar to GP|20805105|dbj|BAB92777. contains ESTs AU065... 95 3e-20
TC86332 similar to GP|7528276|gb|AAF63205.1| AP2-related transcr... 94 3e-20
>TC78754 homologue to GP|4099921|gb|AAD09248.1| EREBP-3 homolog
{Stylosanthes hamata}, partial (46%)
Length = 1038
Score = 274 bits (701), Expect = 2e-74
Identities = 152/220 (69%), Positives = 166/220 (75%)
Frame = +3
Query: 9 AVKVSGNGNGNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAY 68
A KV NG NG VKE HFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAY
Sbjct: 102 APKVVKNGGVNG---VKEAHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAY 272
Query: 69 DTAAREFRGPKAKTNFPFPSDNVKNQSPSQSSTVESSSRDRDVAAAADSSSLDLNLAPSV 128
D+AAR+FRGPKAKTNFPFP +N+K SPSQSSTVESSSRDRD AAADSS LDLNLAP+
Sbjct: 273 DSAARQFRGPKAKTNFPFPIENIKIHSPSQSSTVESSSRDRD--AAADSSPLDLNLAPA- 443
Query: 129 RFPFRPQFPQMPAANQVLFFDAILRAGMVGGSPASQRYGFDFHTYNHPVAASEIQAVAGA 188
F+ +F MPA NQ FD +LRAGMV S + YGF+ YN + AS A GA
Sbjct: 444 ---FQHRFTPMPAVNQTFCFDPVLRAGMV-NSRSCYGYGFE---YNPSMEASGFHATVGA 602
Query: 189 QSDSDSSSVIDLNHYESGVVIKAGGRNFDLDLNYPPMEDM 228
QSDSDSSSVIDLNH+ GR+FD DLNYPP+EDM
Sbjct: 603 QSDSDSSSVIDLNHH---------GRSFDFDLNYPPLEDM 695
>TC76339 homologue to GP|4099914|gb|AAD00708.1| ethylene-responsive element
binding protein homolog {Stylosanthes hamata}, partial
(37%)
Length = 1110
Score = 171 bits (433), Expect = 2e-43
Identities = 119/236 (50%), Positives = 133/236 (55%), Gaps = 17/236 (7%)
Frame = +2
Query: 8 NAVKVSGNGNGNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARA 67
NA + N N E+HFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARA
Sbjct: 80 NAANLKIKKPSNNNNNATEIHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARA 259
Query: 68 YDTAAREFRGPKAKTNFPFPSDNVKNQSPSQSSTVESS--SRDRDVA---AAADSSSLDL 122
YD AAR+FRGPKAKTNFP P +VK SPSQSSTVESS + DR+V S +D
Sbjct: 260 YDNAARQFRGPKAKTNFPPPPSDVKEDSPSQSSTVESSVPAPDREVTRREVPTGGSVMD- 436
Query: 123 NLAPSVRFPFRP-QFPQMPAAN----------QVLFFDAILRAGMVGGSPASQRYGFDFH 171
RFPF Q MP A V F+D RA + +QR+ F
Sbjct: 437 ------RFPFLSIQQQIMPFAGAGAGAVDGMVPVFFYDPAGRAEFL-----NQRFTNRFE 583
Query: 172 TYNHPVAASEIQAVAGAQSDSDSSSVIDLNHYESGVVIKAGGRNFDLDLNY-PPME 226
PV + I G QSDSDSSSV+D R +LDLN PPME
Sbjct: 584 P--EPVQFN-IGFGGGVQSDSDSSSVVDCQ----------PKRALNLDLNLAPPME 712
>TC86273 similar to SP|O80340|ERF4_ARATH Ethylene responsive element binding
factor 4 (AtERF4). [Mouse-ear cress] {Arabidopsis
thaliana}, partial (43%)
Length = 1171
Score = 167 bits (422), Expect = 4e-42
Identities = 119/238 (50%), Positives = 133/238 (55%), Gaps = 9/238 (3%)
Frame = +3
Query: 1 MAPRDK--TNAVKVSGNGNGNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTF 58
MAPRD TN G KE+ +RGVRKRPWGRYAAEIRDPGKK+RVWLGTF
Sbjct: 90 MAPRDNRITNTTTGPGPNPSAQAQAQKEIRYRGVRKRPWGRYAAEIRDPGKKTRVWLGTF 269
Query: 59 DTAEEAARAYDTAAREFRGPKAKTNFPFPSDNVKNQSPSQSSTVESSSRDRDVAAAADSS 118
DTAEEAARAYD AAREFRG KAKTNFP P + + N SPSQSST+ES S
Sbjct: 270 DTAEEAARAYDKAAREFRGSKAKTNFPTPLEIINNGSPSQSSTLESPS----------PP 419
Query: 119 SLDLNLAP------SVRFPFRPQFPQMPAANQVLFFDAILRAGMVGGSPASQRYGFDFHT 172
LDL L P V + P A VLFFDA RA + GF+
Sbjct: 420 PLDLTLTPFSSSHGGVTLAY-------PVARPVLFFDAFARAETALTVGRREMCGFE--- 569
Query: 173 YNHPVAASEIQAVAGAQSDS-DSSSVIDLNHYESGVVIKAGGRNFDLDLNYPPMEDMA 229
P+A AV QSDS SSSV+D YE GV + R DLDLN PP ++A
Sbjct: 570 --RPMADFRRAAV---QSDSGSSSSVVD---YE-GVPRQ---RLLDLDLNVPPPPEVA 707
>AL389598 homologue to GP|4099914|gb|A ethylene-responsive element binding
protein homolog {Stylosanthes hamata}, partial (26%)
Length = 480
Score = 123 bits (308), Expect = 7e-29
Identities = 60/68 (88%), Positives = 62/68 (90%)
Frame = +2
Query: 38 GRYAAEIRDPGKKSRVWLGTFDTAEEAARAYDTAAREFRGPKAKTNFPFPSDNVKNQSPS 97
GRYAAEIRDPGKKSRVWLGTFDTAEEAARAYD AAR+FRGPKAKTNFP P +VK SPS
Sbjct: 236 GRYAAEIRDPGKKSRVWLGTFDTAEEAARAYDNAARQFRGPKAKTNFPPPPSDVKEDSPS 415
Query: 98 QSSTVESS 105
QSSTVESS
Sbjct: 416 QSSTVESS 439
>TC90000 homologue to GP|9294548|dbj|BAB02811.1 ethylene responsive element
binding factor-like protein {Arabidopsis thaliana},
partial (25%)
Length = 458
Score = 114 bits (285), Expect = 3e-26
Identities = 58/92 (63%), Positives = 64/92 (69%), Gaps = 14/92 (15%)
Frame = +2
Query: 8 NAVKVSGNGNGNGNGGV--------------KEVHFRGVRKRPWGRYAAEIRDPGKKSRV 53
N+ K+ G GNG KE +RGVRKRPWGR+AAEIRDP KK+RV
Sbjct: 161 NSKKIEPLKMGRGNGATATTAAEPGSNPVFFKEPRYRGVRKRPWGRFAAEIRDPLKKARV 340
Query: 54 WLGTFDTAEEAARAYDTAAREFRGPKAKTNFP 85
WLGTFDTAE+AARAYDTAAR RGPKAKTNFP
Sbjct: 341 WLGTFDTAEQAARAYDTAARNLRGPKAKTNFP 436
>TC88083 similar to GP|4587373|dbj|BAA76734.1 ethylene responsive element
binding factor {Nicotiana tabacum}, partial (46%)
Length = 1214
Score = 111 bits (277), Expect = 3e-25
Identities = 80/231 (34%), Positives = 115/231 (49%), Gaps = 27/231 (11%)
Frame = +1
Query: 24 VKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAYDTAAREFRGPKAKTN 83
VKE+ +RGVRKRPWGR+AAEIRDP KK+RVWLGT+DTAE+AA AYD+AA +FR KAKTN
Sbjct: 142 VKEIRYRGVRKRPWGRFAAEIRDPWKKTRVWLGTYDTAEQAALAYDSAAIKFRASKAKTN 321
Query: 84 FPFP---------SDNVKNQSPSQSSTVESSSRDRDVAAAADSSSLDLN------LAPSV 128
FP P + V Q+P + V+ +R +V + + +N ++ +V
Sbjct: 322 FPIPEHILAAAAETAAVAAQNPQPPAAVD--NRPAEVLKFVEPDVVQVNWPTSSGMSSTV 495
Query: 129 RFPFRPQFPQMPAANQVLFFDAILRAG---------MVGGSPASQRYGFDFHT---YNHP 176
P+ Q+ ++ + G VGG+ A G DFH+ +
Sbjct: 496 ESFSGPRIVQLVGSSSSSVVSRVPTVGGGAGGGVAQAVGGAGAGA--GEDFHSGCDSSSS 669
Query: 177 VAASEIQAVAGAQSDSDSSSVIDLNHYESGVVIKAGGRNFDLDLNYPPMED 227
V + V + S + H + + A + F +DLN PP D
Sbjct: 670 VVDDDEDCVILSSSAASVRKPPVQAHALAQAQVHAQAQVFAIDLNLPPPMD 822
>TC80833 weakly similar to PIR|D86175|D86175 hypothetical protein [imported]
- Arabidopsis thaliana, partial (60%)
Length = 709
Score = 110 bits (276), Expect = 3e-25
Identities = 54/94 (57%), Positives = 71/94 (75%), Gaps = 1/94 (1%)
Frame = +1
Query: 10 VKVSGNGNGNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKK-SRVWLGTFDTAEEAARAY 68
V S NGNGNGNG ++ +RG+R+RPWG++AAEIRDP +K +R+WLGTFDTAE+AARAY
Sbjct: 121 VSNSTNGNGNGNGNSDQIKYRGIRRRPWGKFAAEIRDPTRKGTRIWLGTFDTAEQAARAY 300
Query: 69 DTAAREFRGPKAKTNFPFPSDNVKNQSPSQSSTV 102
D AA FRG +A NFP + Q+P+ SS++
Sbjct: 301 DAAAFHFRGHRAILNFP-----NEYQAPNSSSSL 387
>TC79935 homologue to GP|15207792|dbj|BAB62912. ERF domain protein12
{Arabidopsis thaliana}, partial (41%)
Length = 878
Score = 107 bits (268), Expect = 3e-24
Identities = 50/61 (81%), Positives = 53/61 (85%)
Frame = +1
Query: 25 KEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAYDTAAREFRGPKAKTNF 84
+E H+RGVRKRPWGRYAAEIRDP KK+RVWLGTFDT EEAA AYD AAR RG KAKTNF
Sbjct: 157 REGHYRGVRKRPWGRYAAEIRDPWKKTRVWLGTFDTPEEAALAYDGAARSLRGAKAKTNF 336
Query: 85 P 85
P
Sbjct: 337 P 339
>AW559374 similar to GP|17381278|gb| AT4g34410/F10M10_180 {Arabidopsis
thaliana}, partial (27%)
Length = 732
Score = 103 bits (256), Expect = 7e-23
Identities = 46/64 (71%), Positives = 55/64 (85%)
Frame = +3
Query: 29 FRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAYDTAAREFRGPKAKTNFPFPS 88
+RGVR+RPWG++AAEIRDP + +RVWLGTF TAEEAARAYD AA EFRGP+AK NFP
Sbjct: 300 YRGVRQRPWGKWAAEIRDPRRATRVWLGTFSTAEEAARAYDNAAIEFRGPRAKLNFPMVD 479
Query: 89 DNVK 92
D++K
Sbjct: 480 DSLK 491
>TC90582 similar to GP|17381278|gb|AAL36057.1 AT4g34410/F10M10_180
{Arabidopsis thaliana}, partial (29%)
Length = 979
Score = 102 bits (253), Expect = 2e-22
Identities = 46/65 (70%), Positives = 55/65 (83%)
Frame = +2
Query: 29 FRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAYDTAAREFRGPKAKTNFPFPS 88
+RGVR+RPWG++AAEIRDP + RVWLGTF TAEEAARAYD AA EFRGP+AK NFP
Sbjct: 341 YRGVRQRPWGKWAAEIRDPRRAVRVWLGTFTTAEEAARAYDNAAIEFRGPRAKLNFPVVD 520
Query: 89 DNVKN 93
+++KN
Sbjct: 521 ESLKN 535
>TC90274 similar to GP|10177219|dbj|BAB10294. contains similarity to AP2
domain transcription factor~gene_id:MPF21.9 {Arabidopsis
thaliana}, partial (32%)
Length = 750
Score = 101 bits (252), Expect = 2e-22
Identities = 65/152 (42%), Positives = 87/152 (56%), Gaps = 17/152 (11%)
Frame = +3
Query: 29 FRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAYDTAAREFRGPKAKTNFPFPS 88
+RGVR+RPWG++AAEIRDP K +RVWLGTF+TAE AARAYD AA FRG KAK NFP
Sbjct: 165 YRGVRQRPWGKWAAEIRDPFKAARVWLGTFETAEAAARAYDEAALRFRGNKAKLNFP--- 335
Query: 89 DNVKNQSPSQSSTVES---SSRDRDVAAAADSSSLDLNLAP-------SVRFPFRPQFPQ 138
+NV ++P ++T S+ + + S+ ++ P S F R QF
Sbjct: 336 ENVTLRNPPPATTTTQWNVSNSPSSIVSITTSTDPVVHTRPFPNSSQHSTNFYDRFQFSG 515
Query: 139 MPAANQVLFFDAILRAGMV-------GGSPAS 163
+P A V + D ++RA + SPAS
Sbjct: 516 IPPARNV-YDDNVIRASSMASHLQSSSSSPAS 608
>TC78789 homologue to PIR|T48580|T48580 hypothetical protein T31B5.150 -
Arabidopsis thaliana, partial (33%)
Length = 1112
Score = 100 bits (249), Expect = 5e-22
Identities = 53/100 (53%), Positives = 69/100 (69%)
Frame = +1
Query: 25 KEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAYDTAAREFRGPKAKTNF 84
K+ H+RGVR+RPWG++AAEIRDP K +RVWLGTFDTAE+AA AYD AA +F+G KAK NF
Sbjct: 346 KKPHYRGVRQRPWGKWAAEIRDPKKAARVWLGTFDTAEDAALAYDKAALKFKGTKAKLNF 525
Query: 85 PFPSDNVKNQSPSQSSTVESSSRDRDVAAAADSSSLDLNL 124
P + V Q S SST ++ + D + + + NL
Sbjct: 526 P---ERVV-QCNSYSSTANNAIQQSDYVSNSHDQQVFPNL 633
>BG586667 similar to GP|17381278|gb| AT4g34410/F10M10_180 {Arabidopsis
thaliana}, partial (28%)
Length = 740
Score = 100 bits (249), Expect = 5e-22
Identities = 45/65 (69%), Positives = 55/65 (84%)
Frame = +3
Query: 29 FRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAYDTAAREFRGPKAKTNFPFPS 88
+RGVR+RPWG++AAEIRDP + RVWLGTF TAEEAARAYD AA EFRGP+AK NFP
Sbjct: 129 YRGVRQRPWGKWAAEIRDPRRAVRVWLGTFTTAEEAARAYDNAAIEFRGPRAKLNFPLVD 308
Query: 89 DNVKN 93
+++K+
Sbjct: 309 ESLKH 323
>TC76517 similar to GP|11994632|dbj|BAB02769. AP2 domain transcription
factor RAP2.3 {Arabidopsis thaliana}, partial (33%)
Length = 1290
Score = 98.6 bits (244), Expect = 2e-21
Identities = 51/118 (43%), Positives = 68/118 (57%)
Frame = +3
Query: 6 KTNAVKVSGNGNGNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAA 65
K + V V G ++ +RG+R+RPWG++AAEIRDP + RVWLGTF TAEEAA
Sbjct: 507 KQSLVTVEKGKKSTGGKRARKNVYRGIRQRPWGKWAAEIRDPQQGVRVWLGTFSTAEEAA 686
Query: 66 RAYDTAAREFRGPKAKTNFPFPSDNVKNQSPSQSSTVESSSRDRDVAAAADSSSLDLN 123
RAYDTAA+ RG KAK N FP P++ V S ++ + S +L+
Sbjct: 687 RAYDTAAKRIRGDKAKLN--FPDTPAVTAPPAKKQCVSSDQASSQASSESAGSGFELD 854
>BF650547 similar to GP|9758388|dbj| AP2 domain transcription factor-like
{Arabidopsis thaliana}, partial (24%)
Length = 418
Score = 98.2 bits (243), Expect = 2e-21
Identities = 49/84 (58%), Positives = 61/84 (72%), Gaps = 5/84 (5%)
Frame = -1
Query: 28 HFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAYDTAAREFRGPKAKTNFP-- 85
H+RGVR+RPWG++AAEIRDP K +RVWLGTF+TAE AA AYD AA F+G KAK NFP
Sbjct: 388 HYRGVRQRPWGKWAAEIRDPKKGARVWLGTFETAEAAATAYDEAALRFKGNKAKLNFPRL 209
Query: 86 ---FPSDNVKNQSPSQSSTVESSS 106
PSD ++ + +S+ SSS
Sbjct: 208 ISNLPSDPKPPRNKNNASSSSSSS 137
>TC76795 similar to GP|18496063|emb|CAD21849. ethylene responsive element
binding protein {Fagus sylvatica}, partial (32%)
Length = 966
Score = 97.4 bits (241), Expect = 4e-21
Identities = 50/103 (48%), Positives = 64/103 (61%), Gaps = 3/103 (2%)
Frame = +3
Query: 4 RDKTNAVKVSGNGNGNGNGGVKEVH---FRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT 60
R + A V+ N G +K +RG+R+RPWG++AAEIRDP K RVWLGTF+T
Sbjct: 282 RGSSAAKVVASKSNEQGEKELKRKRKNQYRGIRQRPWGKWAAEIRDPRKGVRVWLGTFNT 461
Query: 61 AEEAARAYDTAAREFRGPKAKTNFPFPSDNVKNQSPSQSSTVE 103
AEEAARAYD AR RG KAK NFP + N ++ +S +
Sbjct: 462 AEEAARAYDAEARRIRGKKAKVNFPEEAPNASSKRLKTNSETQ 590
>TC93662 similar to GP|10177219|dbj|BAB10294. contains similarity to AP2
domain transcription factor~gene_id:MPF21.9 {Arabidopsis
thaliana}, partial (31%)
Length = 555
Score = 96.3 bits (238), Expect = 9e-21
Identities = 54/115 (46%), Positives = 72/115 (61%)
Frame = +3
Query: 23 GVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAYDTAAREFRGPKAKT 82
G + +RGVR+RPWG++AAEIRDP K +RVWLGTF+TAE AARAYD AA FRG +AK
Sbjct: 240 GERRRRYRGVRQRPWGKWAAEIRDPHKAARVWLGTFETAEAAARAYDEAALRFRGNRAKL 419
Query: 83 NFPFPSDNVKNQSPSQSSTVESSSRDRDVAAAADSSSLDLNLAPSVRFPFRPQFP 137
NFP +NV + + V +S+ ++ S + + AP P + QFP
Sbjct: 420 NFP---ENVPASATRPTFPVNTST-------SSVSPATHYSPAPPQIQPQQQQFP 554
>BF647376 similar to GP|7939533|dbj| Nicotiana EREBP-3-like protein
{Arabidopsis thaliana}, partial (52%)
Length = 457
Score = 95.9 bits (237), Expect = 1e-20
Identities = 52/95 (54%), Positives = 65/95 (67%), Gaps = 2/95 (2%)
Frame = +2
Query: 18 GNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKK-SRVWLGTFDTAEEAARAYDTAAREFR 76
GN N E +RG+R+RPWG++AAEIRDP +K SR+WLGTF+TAE+AARAYD AA FR
Sbjct: 2 GNKNA---EARYRGIRRRPWGKFAAEIRDPTRKGSRIWLGTFETAEQAARAYDAAAFHFR 172
Query: 77 GPKAKTNFPFP-SDNVKNQSPSQSSTVESSSRDRD 110
G KA NFP S + N P+ +ST S + D
Sbjct: 173 GHKAILNFPNDFSHSSINYIPNTTSTSNSYQQQGD 277
>TC86558 similar to GP|20805105|dbj|BAB92777. contains ESTs AU065506(C0784)
AU083422(C0784)~unknown protein, partial (47%)
Length = 1346
Score = 94.7 bits (234), Expect = 3e-20
Identities = 43/61 (70%), Positives = 50/61 (81%)
Frame = +1
Query: 29 FRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAYDTAAREFRGPKAKTNFPFPS 88
+RG+R+RPWG++AAEIRDP K RVWLGTF+TAEEAARAYD AR+ RG KAK NFP
Sbjct: 403 YRGIRQRPWGKWAAEIRDPRKGVRVWLGTFNTAEEAARAYDKEARKIRGKKAKVNFPNED 582
Query: 89 D 89
D
Sbjct: 583 D 585
>TC86332 similar to GP|7528276|gb|AAF63205.1| AP2-related transcription
factor {Mesembryanthemum crystallinum}, partial (49%)
Length = 1349
Score = 94.4 bits (233), Expect = 3e-20
Identities = 50/93 (53%), Positives = 63/93 (66%), Gaps = 10/93 (10%)
Frame = +1
Query: 24 VKEVHFRGVRKRPWGRYAAEIRDPGKK-SRVWLGTFDTAEEAARAYDTAAREFRGPKAKT 82
VK H+RGVR+RPWG++AAEIRDP K +RVWLGTF+TAE+AA AYD AA RG +A
Sbjct: 526 VKGKHYRGVRQRPWGKFAAEIRDPAKNGARVWLGTFETAEDAALAYDRAAYRMRGSRALL 705
Query: 83 NFPFPSDN---------VKNQSPSQSSTVESSS 106
NFP ++ K SP +SS+ ES+S
Sbjct: 706 NFPLRVNSGEPDPVRIASKRSSPERSSSSESNS 804
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.131 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,067,072
Number of Sequences: 36976
Number of extensions: 76009
Number of successful extensions: 598
Number of sequences better than 10.0: 136
Number of HSP's better than 10.0 without gapping: 499
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 553
length of query: 229
length of database: 9,014,727
effective HSP length: 93
effective length of query: 136
effective length of database: 5,575,959
effective search space: 758330424
effective search space used: 758330424
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0301c.1


