

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0299.5
(547 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
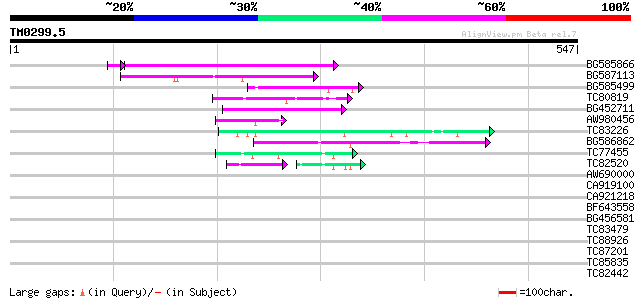
Score E
Sequences producing significant alignments: (bits) Value
BG585866 140 7e-34
BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At... 67 2e-11
BG585499 59 5e-09
TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non... 54 2e-07
BG452711 48 8e-06
AW980456 48 1e-05
TC83226 weakly similar to PIR|G86419|G86419 probable reverse tra... 46 4e-05
BG586862 44 1e-04
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 44 2e-04
TC82520 40 4e-04
AW690000 37 0.025
CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [i... 36 0.043
CA921218 33 0.22
BF643558 33 0.22
BG456581 32 0.63
TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protei... 32 0.82
TC88926 similar to GP|7290507|gb|AAF45960.1| peb gene product {D... 31 1.1
TC87201 similar to PIR|F86388|F86388 hypothetical protein AAG292... 31 1.1
TC85835 similar to PIR|T12180|T12180 probable transcription fact... 31 1.1
TC82442 31 1.4
>BG585866
Length = 828
Score = 140 bits (354), Expect(2) = 7e-34
Identities = 65/208 (31%), Positives = 110/208 (52%), Gaps = 1/208 (0%)
Frame = +3
Query: 111 ILWARDMIDQRFEFRIGKGDTSVWYQDWSGIGIIANQIPFVHISDVNLTLCDLIQDNKWN 170
I+ ++++ + +R G G++S WY +WS +G++ Q PFV I D++LT+ D+ +
Sbjct: 84 IIRDKNVLKSGYTWRAGSGNSSFWYTNWSSLGLLGTQAPFVDIHDLHLTVKDVFTTGGQH 263
Query: 171 LQRLYTNLPHSLQQQFLAVQPQICMNREDAWIWKDGSSGRYSVRDAYEWINHLAHNP-IE 229
Q LYT LP + + + DA+IW S+G Y+ + Y WI
Sbjct: 264 TQSLYTILPTDIAEVINNTHLNFNASIGDAYIWPHNSNGVYTAKSGYSWILSQTETVNYN 443
Query: 230 DRKLNWVWKLRVPEKIRMFTWQVLHNAIPVNELWVRCHLASDATCARCGNVVEDGLHCLR 289
+ +W+W+L++PEK + F W HNA+P L ++ + A C+RCG E HC+R
Sbjct: 444 NSSWSWIWRLKIPEKYKFFLWLACHNAVPTLSLLNHRNMVNSAICSRCGEHEESFFHCVR 623
Query: 290 DCSFS*DLWRRMGAINWRNFRYNNIISW 317
DC FS +W ++G + F +++ W
Sbjct: 624 DCRFSKIIWHKIGFSSPDFFSSSSVQDW 707
Score = 21.6 bits (44), Expect(2) = 7e-34
Identities = 7/17 (41%), Positives = 10/17 (58%)
Frame = +1
Query: 95 IHKVQAHQHDSPIWKGI 111
+H AH++ SP W I
Sbjct: 34 LHIAAAHRNSSPSWSSI 84
>BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (10%)
Length = 767
Score = 66.6 bits (161), Expect = 2e-11
Identities = 52/210 (24%), Positives = 89/210 (41%), Gaps = 19/210 (9%)
Frame = -3
Query: 108 WKGILWARDMIDQRFEFRIGKGD-TSVWYQDWSGIGIIANQIPFVHISDVN--LTL---- 160
W+ I A+ +I Q + IG G+ T++W ++ + P H S TL
Sbjct: 726 WRSIHSAQHLIKQGAKVIIGNGENTNIWEREMAWKLTCVTNHPNKHSSRAY*APTLYGYE 547
Query: 161 ---CDLIQDNKWNLQRLYTNLPHSLQQQFLAVQPQICMNREDAWIWKDGSSGRYSVRDAY 217
D + N + + P +++ L++ PQ + ED++ W+ SG YSV+ Y
Sbjct: 546 GCRSDDPMRRERNANLINSIFPEGTRRKILSIHPQGPIG-EDSYSWEYSKSGHYSVKSGY 370
Query: 218 EWINHL---------AHNPIEDRKLNWVWKLRVPEKIRMFTWQVLHNAIPVNELWVRCHL 268
++ P D VWK K+R F W+ + N++P H+
Sbjct: 369 YVQTNIIAAANQRGTVDQPSLDDLYQRVWKYNTSPKVRHFLWRCISNSLPTAANMRSRHI 190
Query: 269 ASDATCARCGNVVEDGLHCLRDCSFS*DLW 298
+ D +C+RCG E H L C ++ +W
Sbjct: 189 SKDGSCSRCGMESETVNHILFQCPYARLIW 100
>BG585499
Length = 792
Score = 58.9 bits (141), Expect = 5e-09
Identities = 37/122 (30%), Positives = 54/122 (43%), Gaps = 10/122 (8%)
Frame = +3
Query: 230 DRKLNWVWKLRVPEKIRMFTWQVLHNAIPVNELWVRCHLASDATCARCGNVVEDGLHCLR 289
D K+ W W R P + + F W V H I N R ATC CGN E LH L
Sbjct: 222 DWKMLWGW--RGPHRTQTFMWLVAHGCILTNYRRSRWGTRVLATCPCCGNADETVLHVLC 395
Query: 290 DCSFS*DLWRRMGAINW--RNFRYNNIISW-FSSMARGVHGIQ-------FLAGVWGAWK 339
DC + +W R+ +W F +++ W F ++++ +G+ F+ W W
Sbjct: 396 DCRPASQVWIRLVPSDWITNFFSFDDCRDWVFKNLSKRSNGVSKFKWQPTFMTTCWHMWT 575
Query: 340 WR 341
WR
Sbjct: 576 WR 581
>TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non-LTR
retroelement reverse transcriptase {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 1262
Score = 53.5 bits (127), Expect = 2e-07
Identities = 45/139 (32%), Positives = 59/139 (42%), Gaps = 4/139 (2%)
Frame = +2
Query: 196 NREDAWIWKDGSSGRYSVRDAYEWINHLAHNPIEDRKL-NWVWKLRVPEKIRMFTWQVLH 254
N D W W YSV+ Y +I H I DR L + VW +P K+ +F W++L
Sbjct: 509 NVNDKWRWLLDPVNGYSVKVFYRYITSTGH--ISDRSLVDDVWHKHIPSKVSLFVWRLLR 682
Query: 255 NAIPVNELWVR--CHLASDATCARCGNV-VEDGLHCLRDCSFS*DLWRRMGAINWRNFRY 311
N +P + V LA++A C CG V E H C+ LW + NW
Sbjct: 683 NRLPTKDNLVHRGVLLATNAACV-CGCVDSESTTHLFLHCNVFCSLWSLVR--NWLG--- 844
Query: 312 NNIISWFSSMARGVHGIQF 330
I SS H IQF
Sbjct: 845 ---IPSMSSGELRTHFIQF 892
>BG452711
Length = 672
Score = 48.1 bits (113), Expect = 8e-06
Identities = 27/120 (22%), Positives = 51/120 (42%)
Frame = +3
Query: 206 GSSGRYSVRDAYEWINHLAHNPIEDRKLNWVWKLRVPEKIRMFTWQVLHNAIPVNELWVR 265
GS+ V Y W+ + + + NW+ +L P I++F Q+ + + + +
Sbjct: 204 GSTAHLIVSKGYWWLMGVHTSFLGKES*NWISRLCAPSNIKLFL*QL*RDYVHFRSILLF 383
Query: 266 CHLASDATCARCGNVVEDGLHCLRDCSFS*DLWRRMGAINWRNFRYNNIISWFSSMARGV 325
C+L S C C +D LH L C+ + D+W + +N+I+W +
Sbjct: 384 CNLISSNLCPICNQRSQDMLHALFSCTRAKDVWSFCLHELVTSSNQDNLITWMKEFISNI 563
>AW980456
Length = 779
Score = 47.8 bits (112), Expect = 1e-05
Identities = 24/72 (33%), Positives = 37/72 (51%), Gaps = 3/72 (4%)
Frame = -2
Query: 199 DAWIWKDGSSGRYSVRDAYEW-INHLAHNPIEDRKLNW--VWKLRVPEKIRMFTWQVLHN 255
D IWKD G+Y V+ AY + + L + R NW +WKL+VP K++ W++
Sbjct: 208 DRLIWKDEKHGKYYVKSAYRFCVEELFDSSYLHRPGNWSGIWKLKVPPKVQNLVWRMCRG 29
Query: 256 AIPVNELWVRCH 267
+P +R H
Sbjct: 28 CLPTR---IRLH 2
>TC83226 weakly similar to PIR|G86419|G86419 probable reverse transcriptase
100033-105622 [imported] - Arabidopsis thaliana, partial
(2%)
Length = 885
Score = 45.8 bits (107), Expect = 4e-05
Identities = 66/295 (22%), Positives = 112/295 (37%), Gaps = 29/295 (9%)
Frame = +3
Query: 202 IWKDGSSGRYSVRDAYE----WINHLAHNPI--EDRKLNW--VWKLRVPEKIRMFTWQVL 253
+W +G YSV+ Y W +N D L W +W L + ++ W++L
Sbjct: 3 MWMHNPTGIYSVKSGYNTLRTWQTQQINNTSTSSDETLIWKKIWSLHTIPRHKVLLWRIL 182
Query: 254 HNAIPVNELWVRCHLASDATCARCGNVVEDGLHCLRDCSFS*DLWRRMG-AINWRNFRYN 312
++++PV + + C RC + E H C S +W IN+ N
Sbjct: 183 NDSLPVRSSLRKRGIQCYPLCPRCHSKTETITHLFMSCPLSKRVWFGSNLCINFDNLPNP 362
Query: 313 NIISWFSSM---ARGVHGIQFLAGVWGAWKWRCNWLLDSQRWPIEVVWRRIAHDHDDW-- 367
N I+ I A ++ W R +L+ Q + +R ++ D+
Sbjct: 363 NFIN*LYEAIL*KDECITI*IAAIIYNLWHARNLSVLEDQTILEMDIIQRASNCISDYKQ 542
Query: 368 --AWCAPSNDLLLCHP-----------WSPPPPDTVKCNSDGSFREDVQRMGGVGVIRDH 414
PS P W P VK N+D + + + G+ +IRD
Sbjct: 543 ANTQAPPSMARTGYDPRSQHRPAKNTKWKRPNLGLVKVNTDANLQNHGKWGLGI-IIRDE 719
Query: 415 QGRWVAGCYLGEAAGN--AFRAEAKALLDVLELAWNRGYSRLICDVNCDNLVTIL 467
G V E GN A AEA ALL + A + G+ ++ + + + L+ ++
Sbjct: 720 VG-LVMAASTWETDGNDRALEAEAYALLTGMRFAKDCGFXKVXFEGDNEKLMKMV 881
>BG586862
Length = 804
Score = 44.3 bits (103), Expect = 1e-04
Identities = 49/237 (20%), Positives = 98/237 (40%), Gaps = 8/237 (3%)
Frame = -1
Query: 236 VWKLRVPEKIRMFTWQVLHNAIPVNELWVRCHLASDATCARCGNVVEDGLHCLRDCSFS* 295
VW ++ + + F W++LHNA+PV + + + C RC + +E H +C +
Sbjct: 654 VWGIKTIPRHKSFLWRLLHNALPVKDELHKRGIRCSLLCPRCESKIETVQHLFLNCEVTQ 475
Query: 296 DLWRRMGAINWRNFRYNNIISWFSSMARGVHG------IQFLAGVWGAWKWRCNWLLDSQ 349
W G+ NF + ++ + + + I A ++ W R + ++
Sbjct: 474 KEW--FGSQLGINFHSSGVLHFHDWITNFILKNDEETIIALTALLYSIWHARNQKVFENI 301
Query: 350 RWPIEVVWRRIAHDHDDWAWCAPSNDLLLCHPWSPPPPDTVKCNSDGSFREDVQRMGGVG 409
P +VV +R + + S+ +L P + + S + G+G
Sbjct: 300 DVPGDVVIQRASSSLHSFKMAQVSDSVL--------PSNAIPSYS----------LWGIG 175
Query: 410 VI-RDHQGRWVA-GCYLGEAAGNAFRAEAKALLDVLELAWNRGYSRLICDVNCDNLV 464
V+ R+ +G +A G +L A AEA + + A + G+S+ + + NL+
Sbjct: 174 VVARNCEGLAMASGTWLRHGIPCATTAEAWGIYQAMVFAGDCGFSKFEFESDNGNLL 4
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 43.5 bits (101), Expect = 2e-04
Identities = 39/150 (26%), Positives = 59/150 (39%), Gaps = 13/150 (8%)
Frame = -2
Query: 199 DAWIWKDGSSGRYSVRDAYEWINHLAHNPIEDRK-------LNWVWKLRVPEKIRMFTWQ 251
D W+WK G +SV Y + +L +EDR +WK + P K+ F+W
Sbjct: 448 DIWVWKPDKEGVFSVNSCYFLLQNL--RLLEDRLSYEEEVIFRELWKSKAPAKVLAFSWT 275
Query: 252 VLHNAIP--VNELWVRCHLASDA-TCARCGNVVEDGLHCLRDCSFS*DLWRRMGAINWRN 308
+ + IP VN R D+ C CG E +H C + R + + W N
Sbjct: 274 LFLDRIPTMVNLGKRRLLRVEDSKRCVFCGCQDETVVHLFLHCDVISKV*REV--MRWLN 101
Query: 309 FRY---NNIISWFSSMARGVHGIQFLAGVW 335
F N++ +R V + G W
Sbjct: 100 FNLISPPNLLIHAICWSREVRAKKLRQGAW 11
>TC82520
Length = 833
Score = 39.7 bits (91), Expect(2) = 4e-04
Identities = 15/59 (25%), Positives = 34/59 (57%)
Frame = +3
Query: 210 RYSVRDAYEWINHLAHNPIEDRKLNWVWKLRVPEKIRMFTWQVLHNAIPVNELWVRCHL 268
R + R Y ++ ++ P++ +++ VW+ +P K+ MF W++ HN +P ++ H+
Sbjct: 111 RVTRRGTYRFLT-ISGQPLDRNQVDDVWQKNIPSKVSMFVWRLFHNRLPTKVNLMQRHV 284
Score = 21.9 bits (45), Expect(2) = 4e-04
Identities = 22/79 (27%), Positives = 28/79 (34%), Gaps = 12/79 (15%)
Frame = +2
Query: 277 CGNVVEDGLHCLRDCSFS*DLWRRMGAINWRNFRY------NNIISWFSSMA---RGVHG 327
CG+ E H C LW + + W + F+SMA R H
Sbjct: 317 CGDS-ETATHLFLHCDIFGSLWSHV--LRWLHLLLVLPADIRQFFIQFTSMAGSPRFTHS 487
Query: 328 ---IQFLAGVWGAWKWRCN 343
I + A VW WK R N
Sbjct: 488 FLQIMWFASVWVLWKKRNN 544
>AW690000
Length = 652
Score = 36.6 bits (83), Expect = 0.025
Identities = 20/54 (37%), Positives = 25/54 (46%)
Frame = +3
Query: 382 WSPPPPDTVKCNSDGSFREDVQRMGGVGVIRDHQGRWVAGCYLGEAAGNAFRAE 435
W PP P +KCN+DGS R G G+ R+H + NAF AE
Sbjct: 411 WRPPIPHWIKCNTDGSSRSHSSACG--GIFRNHDTDLLLCFAENTGECNAFHAE 566
>CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [imported] -
Sulfolobus solfataricus, partial (3%)
Length = 789
Score = 35.8 bits (81), Expect = 0.043
Identities = 28/124 (22%), Positives = 52/124 (41%), Gaps = 9/124 (7%)
Frame = -2
Query: 236 VWKLRVPEKIRMFTWQVLHNAIPV-NELWVRCHLASDA-TCARCGNVVEDGLHCLRDCSF 293
+W +VP K+ +F W++L + +P + L R + ++A C +E H CS+
Sbjct: 584 IWHRQVPLKVSVFAWRLLRDRLPTKSNLIYRGVIPTEAGLCVSGCGALESAQHLFLSCSY 405
Query: 294 S*DLWRRMGAINW-------RNFRYNNIISWFSSMARGVHGIQFLAGVWGAWKWRCNWLL 346
LW + +W N ++ + + S FL +W C W+L
Sbjct: 404 FASLWSLVR--DWIGFVGVDTNVLSDHFVQFVHSTGGNKASQSFLQLIW----LLCAWVL 243
Query: 347 DSQR 350
++R
Sbjct: 242 WTER 231
>CA921218
Length = 707
Score = 33.5 bits (75), Expect = 0.22
Identities = 30/131 (22%), Positives = 56/131 (41%)
Frame = -3
Query: 385 PPPDTVKCNSDGSFREDVQRMGGVGVIRDHQGRWVAGCYLGEAAGNAFRAEAKALLDVLE 444
P P + CN++GS + G G R+ ++ GNA A+ + +E
Sbjct: 594 PEPLWINCNTNGSANINTSACG--GTFRNSNAYFLLCFAENTGNGNALHAKLSGAMRAIE 421
Query: 445 LAWNRGYSRLICDVNCDNLVTILVEAEAVQMHSEFHVLHSITQLLARDWHVRINSVHRDS 504
+A R +S L +++ +LV ++++V +H + L + ++ V R+
Sbjct: 420 IAAARNWSHLWLELD-SSLVVNAFKSKSV-IHWNLRNRWNNCLFLISSMNFFVSHVFREG 247
Query: 505 NAVADHLVRRG 515
N AD L G
Sbjct: 246 NQCADGLANFG 214
>BF643558
Length = 645
Score = 33.5 bits (75), Expect = 0.22
Identities = 12/26 (46%), Positives = 16/26 (61%)
Frame = +2
Query: 382 WSPPPPDTVKCNSDGSFREDVQRMGG 407
W+PPP +KCN+DGS + GG
Sbjct: 557 WNPPPLHWIKCNTDGSSNTNTSSCGG 634
>BG456581
Length = 683
Score = 32.0 bits (71), Expect = 0.63
Identities = 43/201 (21%), Positives = 70/201 (34%), Gaps = 29/201 (14%)
Frame = +2
Query: 236 VWKLRVPEKIRMFTWQVLHNAIPVNELWVRCHLASDATCARCGNVVEDGLHCLRDCSFS* 295
+W +P W++ H+ +P ++ A + C+ C VE H C F
Sbjct: 80 IWNSCIPPSHSFICWRLAHDRLPTDDNLSSRGCALVSMCSFCLEQVETSDHLFLRCKFVV 259
Query: 296 DLWR------RMGAINWRNFR--YNNIISWFSSMARGVHGIQFLAGVWGAWKWRCNWLLD 347
LW R+G +++ +F+ +++ SS R ++ + V W R N
Sbjct: 260 TLWSWLCSQLRVG-LDFSSFKALLSSLPRHCSSQVRDLYVAAVVHMVHSIWWARNNVRFS 436
Query: 348 SQRWPIEVVWRRI---------AHDHDDWAWCAPSNDLLLCHP------------WSPPP 386
S + V R+ A A D+ P W PP
Sbjct: 437 SAKVSAHAVQVRVHALIGLSGAVSTGKCIAADAAILDVFRIPPHRRSMXEMVSVCWKPPS 616
Query: 387 PDTVKCNSDGSFREDVQRMGG 407
VK N+DGS + GG
Sbjct: 617 APWVKGNTDGSXLNNSGAXGG 679
>TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protein {Plasmodium
falciparum 3D7}, partial (0%)
Length = 1222
Score = 31.6 bits (70), Expect = 0.82
Identities = 21/78 (26%), Positives = 34/78 (42%)
Frame = +2
Query: 373 SNDLLLCHPWSPPPPDTVKCNSDGSFREDVQRMGGVGVIRDHQGRWVAGCYLGEAAGNAF 432
S ++ C W P K N+DGS ++ G G++RD++G + G+ F
Sbjct: 953 SRSIIWCE-WKKPEIGWTKLNTDGSVNKETAGFG--GLLRDYRGEPICAFVSKAPQGDTF 1123
Query: 433 RAEAKALLDVLELAWNRG 450
E A+ L L+ G
Sbjct: 1124 LVELWAIWRGLVLSLGLG 1177
>TC88926 similar to GP|7290507|gb|AAF45960.1| peb gene product {Drosophila
melanogaster}, partial (1%)
Length = 1073
Score = 31.2 bits (69), Expect = 1.1
Identities = 36/132 (27%), Positives = 57/132 (42%), Gaps = 5/132 (3%)
Frame = +3
Query: 388 DTVKCNSDGSF--REDVQRMGGVGVIRDHQGRWVAGCYLGEAAGN--AFRAEAKALLDVL 443
+ V N DGS +V G GV+ D G+W+ G + + N E +A+L L
Sbjct: 336 EEVILNVDGSLLREREVPSAGCGGVLSDSSGKWLCG-FAQKLNPNLKVDETEKEAILRGL 512
Query: 444 ELAWNRGYSRLICDVNCDNLVTILVEAEAVQMHSEFHVLHSITQLL-ARDWHVRINSVHR 502
+G +++ V DN +V + S ++ I LL + W + +H
Sbjct: 513 LWVKEKGKRKIL--VKSDN--EGVVYSVNCGGRSNDPLVCGIRDLLNSPHWEATLTCIHG 680
Query: 503 DSNAVADHLVRR 514
SNAVAD L +
Sbjct: 681 RSNAVADRLAHK 716
>TC87201 similar to PIR|F86388|F86388 hypothetical protein AAG29221.1
[imported] - Arabidopsis thaliana, partial (41%)
Length = 991
Score = 31.2 bits (69), Expect = 1.1
Identities = 22/67 (32%), Positives = 29/67 (42%), Gaps = 4/67 (5%)
Frame = -2
Query: 297 LWRRMGAIN-WRNFRYNNIISWFSSMARGVHGIQFLAGVWGAWKWRCNW---LLDSQRWP 352
LWRR +N WR +R I W+ L +W W WR W L+D RW
Sbjct: 444 LWRRR*FVNIWRRWR*LVDIWWWRWR---------LVYIWWGW*WRW-WRRRLVDIWRWG 295
Query: 353 IEVVWRR 359
+ + W R
Sbjct: 294 VHIWWGR 274
>TC85835 similar to PIR|T12180|T12180 probable transcription factor - fava
bean, partial (79%)
Length = 1737
Score = 31.2 bits (69), Expect = 1.1
Identities = 23/96 (23%), Positives = 31/96 (31%), Gaps = 18/96 (18%)
Frame = +2
Query: 305 NWRNFRYNNIISWFSSMARGVHGIQFLAGVWGAWKWRCNW-----------------LLD 347
NW ++ +S G G+Q A WG W+W W L+
Sbjct: 344 NWCCQEARSVTFQTTSPFSGCEGVQK*AISWGPWRWTWIWTWSR*WWLSS*FLQ**QLIA 523
Query: 348 SQRWPIEVVWRRIAHDH-DDWAWCAPSNDLLLCHPW 382
RR H + W WC P + L C W
Sbjct: 524 CSSCQSRFF*RRFCKSH*ETWVWCTPYS--LSCWRW 625
>TC82442
Length = 578
Score = 30.8 bits (68), Expect = 1.4
Identities = 18/64 (28%), Positives = 28/64 (43%), Gaps = 14/64 (21%)
Frame = -3
Query: 415 QGRWVAGCYLGEAAGNAF--------------RAEAKALLDVLELAWNRGYSRLICDVNC 460
QGRW GC + +AG A AEA A+L + LA +I + +C
Sbjct: 333 QGRWGLGCIIHNSAGQAMTSATWSRNGFNCAATAEAYAILAAMHLALTSDLKHMIFESDC 154
Query: 461 DNLV 464
+ ++
Sbjct: 153 EAVI 142
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.332 0.143 0.506
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,205,315
Number of Sequences: 36976
Number of extensions: 434776
Number of successful extensions: 2664
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 2591
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2654
length of query: 547
length of database: 9,014,727
effective HSP length: 101
effective length of query: 446
effective length of database: 5,280,151
effective search space: 2354947346
effective search space used: 2354947346
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0299.5


